3HVO
 
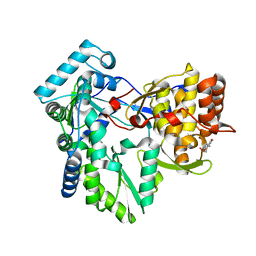 | | Structure of the genotype 2B HCV polymerase bound to a NNI | | Descriptor: | 2-(3-bromophenyl)-6-[(2-hydroxyethyl)amino]-1h-benzo[de]isoquinoline-1,3(2h)-dione, Genome polyprotein | | Authors: | Rydberg, E.H, Carfi, A. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Biological Evaluation of a Series of 1H-Benzo[de]isoquinoline-1,3(2H)-diones as Hepatitis C Virus NS5B Polymerase Inhibitors
To be Published
|
|
2XWY
 
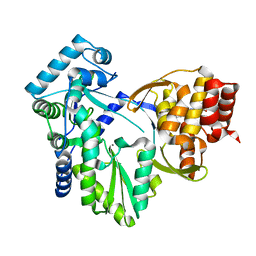 | | Structure of MK-3281, a Potent Non-Nucleoside Finger-Loop Inhibitor, in complex with the Hepatitis C Virus NS5B Polymerase | | Descriptor: | (7R)-14-cyclohexyl-7-{[2-(dimethylamino)ethyl](methyl)amino}-7,8-dihydro-6H-indolo[1,2-e][1,5]benzoxazocine-11-carboxylic acid, MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Di Marco, S, Baiocco, P. | | Deposit date: | 2010-11-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of (7R)-14-Cyclohexyl-7-{[2-(Dimethylamino)Ethyl] (Methyl)Amino}-7,8-Dihydro-6H-Indolo[1,2-E][1,5] Benzoxazocine -11-Carboxylic Acid (Mk-3281), a Potent and Orally Bioavailable Finger-Loop Inhibitor of the Hepatitis C Virus Ns5B Polymerase
J.Med.Chem., 54, 2011
|
|
2WHO
 
 | |
1OL0
 
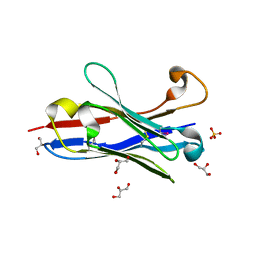 | | Crystal structure of a camelised human VH | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN G, SULFATE ION | | Authors: | Dottorini, T, Vaughan, C.K, Walsh, M.A, Losurdo, P, Sollazzo, M. | | Deposit date: | 2003-08-02 | | Release date: | 2004-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Human Vh: Requirements for Maintaining a Monomeric Fragment
Biochemistry, 43, 2004
|
|
6WCR
 
 | |
3GSZ
 
 | | Structure of the genotype 2B HCV polymerase | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Rydberg, E.H, Carfi, A. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for resistance of the genotype 2b hepatitis C virus NS5B polymerase to site A non-nucleoside inhibitors.
J.Mol.Biol., 390, 2009
|
|
7DBJ
 
 | | Crystal structure of human LDHB in complex with NADH, oxamate, and AXKO-0046 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase B chain, ... | | Authors: | Sogabe, S, Miwa, M. | | Deposit date: | 2020-10-20 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Identification of the first highly selective inhibitor of human lactate dehydrogenase B.
Sci Rep, 11, 2021
|
|
7DBK
 
 | | Crystal structure of human LDHB in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-lactate dehydrogenase B chain | | Authors: | Sogabe, S, Miwa, M. | | Deposit date: | 2020-10-20 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Identification of the first highly selective inhibitor of human lactate dehydrogenase B.
Sci Rep, 11, 2021
|
|
8E1C
 
 | |
8E1B
 
 | |
8E2N
 
 | |
8EE2
 
 | |
8EVD
 
 | |
8F6U
 
 | |
8F6V
 
 | |
8GJR
 
 | |
5TNJ
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 4-Vinyl-1-cyclohexene 1,2-epoxide hydrolysis intermediate | | Descriptor: | (1R,2R,4R)-4-ethenylcyclohexane-1,2-diol, (1R,2R,4S)-4-ethenylcyclohexane-1,2-diol, (1S,2S,4R)-4-ethenylcyclohexane-1,2-diol, ... | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNK
 
 | |
5TNL
 
 | |
5TND
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 1,2-Epoxycyclohexane hydrolysis intermediate | | Descriptor: | (1R,2R)-cyclohexane-1,2-diol, (1S,2S)-cyclohexane-1,2-diol, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNE
 
 | |
5TNM
 
 | |
5TNI
 
 | |
5TNG
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 14,15-EpETE hydrolysis intermediate | | Descriptor: | (5Z,8Z,11Z,14R,15R,17Z)-14,15-dihydroxyicosa-5,8,11,17-tetraenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNP
 
 | |
