4O59
 
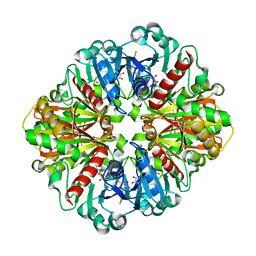 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | Deposit date: | 2013-12-19 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4O63
 
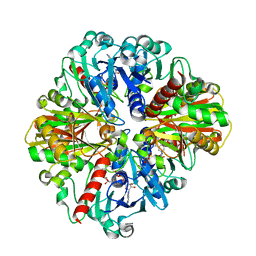 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4OU9
 
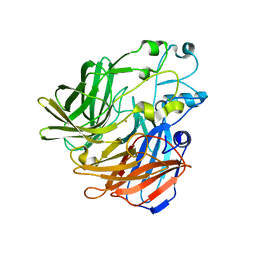 | | Crystal structure of apocarotenoid oxygenase in the presence of Triton X-100 | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Palczewski, K, Kiser, P.D. | | Deposit date: | 2014-02-15 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of Carotenoid Isomerase Activity in a Prototypical Carotenoid Cleavage Enzyme, Apocarotenoid Oxygenase (ACO).
J.Biol.Chem., 289, 2014
|
|
4Q95
 
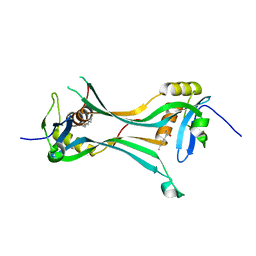 | | Crystal structure of HRASLS3/LRAT chimeric protein | | Descriptor: | HEPTANOIC ACID, HRAS-like suppressor 3, Lecithin retinol acyltransferase | | Authors: | Golczak, M, Kiser, P.D, Sears, A.E, Palczewski, K. | | Deposit date: | 2014-04-29 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | LRAT-specific domain facilitates vitamin A metabolism by domain swapping in HRASLS3.
Nat.Chem.Biol., 11, 2015
|
|
4OU8
 
 | | Crystal structure of apocarotenoid oxygenase in the presence of C8E6 | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Shi, W, Palczewski, K, Kiser, P.D. | | Deposit date: | 2014-02-15 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Carotenoid Isomerase Activity in a Prototypical Carotenoid Cleavage Enzyme, Apocarotenoid Oxygenase (ACO).
J.Biol.Chem., 289, 2014
|
|
4Q2R
 
 | |
4DOT
 
 | | Crystal structure of human HRASLS3. | | Descriptor: | Group XVI phospholipase A2 | | Authors: | Kiser, P.D, Golczak, M, Sears, A.E, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2012-02-10 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for the Acyltransferase Activity of Lecithin:Retinol Acyltransferase-like Proteins.
J.Biol.Chem., 287, 2012
|
|
4DPZ
 
 | | Crystal structure of human HRASLS2 | | Descriptor: | HRAS-like suppressor 2 | | Authors: | Kiser, P.D, Golczak, M, Sears, A.E, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2012-02-14 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Acyltransferase Activity of Lecithin:Retinol Acyltransferase-like Proteins.
J.Biol.Chem., 287, 2012
|
|
4F3D
 
 | |
4F2Z
 
 | |
4F3A
 
 | |
4F3J
 
 | |
4F30
 
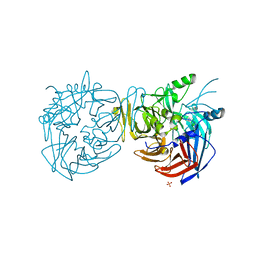 | |
7JTI
 
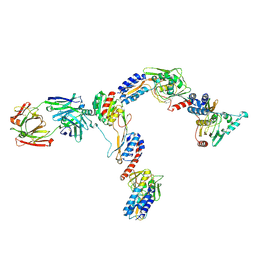 | | Interphotoreceptor retinoid-binding protein (IRBP) in complex with a monoclonal antibody (F3F5 mAb5) | | Descriptor: | Retinol-binding protein 3, mAb5 Fab heavy chain, mAb5 Fab light chain | | Authors: | Sears, A.E, Albiez, S, Gulati, S, Wang, B, Kiser, P, Kovacik, L, Engel, A, Stahlberg, H, Palczewski, K. | | Deposit date: | 2020-08-17 | | Release date: | 2020-10-07 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Single particle cryo-EM of the complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody.
Faseb J., 34, 2020
|
|
7F1W
 
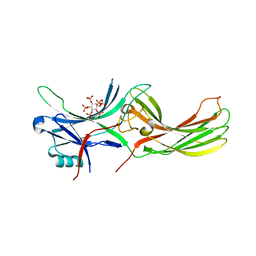 | | X-ray crystal structure of visual arrestin complexed with inositol hexaphosphate | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, S-arrestin | | Authors: | Kang, M, Jang, K, Eger, B.T, Ernst, O.P, Choe, H.W, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7F1X
 
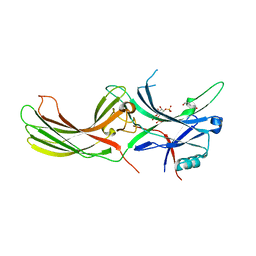 | | X-ray crystal structure of visual arrestin complexed with inositol 1,4,5-triphosphate | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PENTANEDIAL, ... | | Authors: | Jang, K, Kang, M, Eger, B.T, Choe, H.W, Ernst, O.P, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
2R2I
 
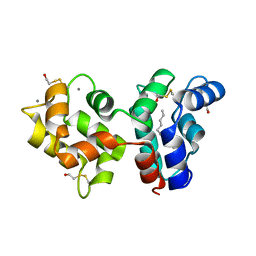 | | Myristoylated Guanylate Cyclase Activating Protein-1 with Calcium Bound | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Guanylyl cyclase-activating protein 1, ... | | Authors: | Stephen, R. | | Deposit date: | 2007-08-25 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1.
Structure, 15, 2007
|
|
3QC9
 
 | |
2GGZ
 
 | |
5ILO
 
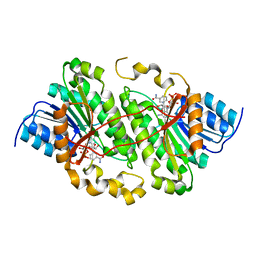 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Photoreceptor dehydrogenase, isoform C | | Authors: | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | Deposit date: | 2016-03-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
5ILG
 
 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | Deposit date: | 2016-03-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
3MZS
 
 | | Crystal Structure of Cytochrome P450 CYP11A1 in complex with 22-hydroxy-cholesterol | | Descriptor: | (3alpha,8alpha,22R)-cholest-5-ene-3,22-diol, Cholesterol side-chain cleavage enzyme, ISOPROPYL ALCOHOL, ... | | Authors: | Stout, C.D, Annalora, A, Mast, N, Pikuleva, I. | | Deposit date: | 2010-05-12 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Three-step Sequential Catalysis by the Cholesterol Side Chain Cleavage Enzyme CYP11A1.
J.Biol.Chem., 286, 2011
|
|
4DG1
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with polymorphism mutation K172A and K173A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tu, X, Kirby, K.A, Marchand, B, Sarafianos, S.G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | HIV-1 Reverse Transcriptase (RT) Polymorphism 172K Suppresses the Effect of Clinically Relevant Drug Resistance Mutations to Both Nucleoside and Non-nucleoside RT Inhibitors.
J.Biol.Chem., 287, 2012
|
|
6C7K
 
 | | Crystal structure of an ACO/RPE65 chimera | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Kiser, P.D, Shi, W. | | Deposit date: | 2018-01-22 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the pathogenesis of dominant retinitis pigmentosa associated with a D477G mutation in RPE65.
Hum.Mol.Genet., 27, 2018
|
|
6C7P
 
 | |
