1POG
 
 | | SOLUTION STRUCTURE OF THE OCT-1 POU-HOMEO DOMAIN DETERMINED BY NMR AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | OCT-1 POU HOMEODOMAIN DNA-BINDING PROTEIN | | Authors: | Cox, M, Van Tilborg, P.J.A, De Laat, W, Boelens, R, Van Leeuwen, H.C, Van Der Vliet, P.C, Kaptein, R. | | Deposit date: | 1994-10-12 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Oct-1 POU homeodomain determined by NMR and restrained molecular dynamics.
J.Biomol.NMR, 6, 1995
|
|
5AI1
 
 | | Crystal structure of ketosteroid isomerase containing Y32F, D40N, Y57F and Y119F mutations in the equilenin-bound form | | Descriptor: | EQUILENIN, KETOSTEROID ISOMERASE | | Authors: | Cha, H.J, Jeong, J.H, Kim, Y.G. | | Deposit date: | 2015-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Contribution of a Low-Barrier Hydrogen Bond to Catalysis is not Significant in Ketosteroid Isomerase.
Mol.Cells, 38, 2015
|
|
8UY4
 
 | | Acinetobacter baumannii Tse15 Rhs effector | | Descriptor: | Tse15, Tse15 toxin peptide (polyUNK) | | Authors: | Hayes, B.K, Venugopal, H, McGowan, S. | | Deposit date: | 2023-11-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of a Rhs effector clade domain provides mechanistic insights into type VI secretion system toxin delivery.
Nat Commun, 15, 2024
|
|
8UXT
 
 | |
7DJT
 
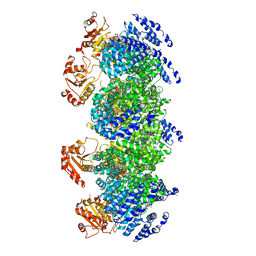 | | Human SARM1 inhibitory state bounded with inhibitor dHNN | | Descriptor: | NAD(+) hydrolase SARM1, O3-methyl O5-(2-methylpropyl) 2,6-dimethyl-4-[2-(oxidanylamino)phenyl]pyridine-3,5-dicarboxylate | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2020-11-21 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Permeant fluorescent probes visualize the activation of SARM1 and uncover an anti-neurodegenerative drug candidate.
Elife, 10, 2021
|
|
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5F1K
 
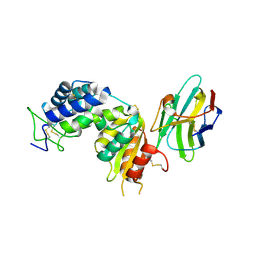 | | human CD38 in complex with nanobody MU1053 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, nanobody MU1053 | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5F1O
 
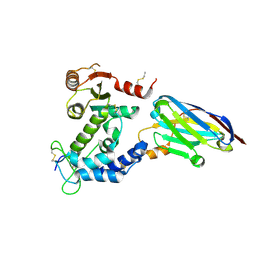 | | human CD38 in complex with nanobody MU551 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, nanobody MU551 | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
3GQG
 
 | | Crystal structure at acidic pH of the ferric form of the Root effect hemoglobin from Trematomus bernacchii. | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vergara, A, Franzese, M, Merlino, A, Bonomi, G, Mazzarella, L. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Correlation between hemichrome stability and the root effect in tetrameric hemoglobins.
Biophys.J., 97, 2009
|
|
8GNJ
 
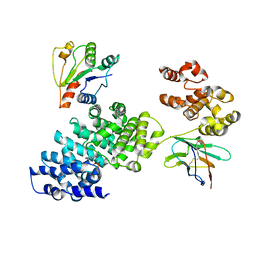 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody-C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8GQ5
 
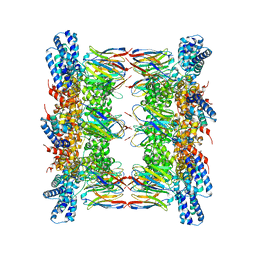 | |
8GNI
 
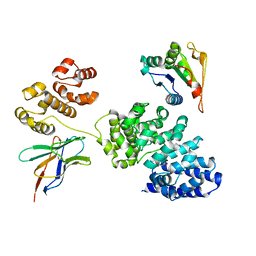 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
2PEG
 
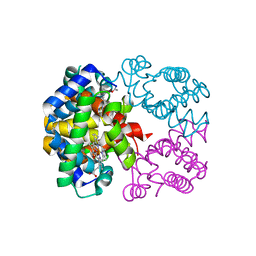 | | Crystal structure of Trematomus bernacchii hemoglobin in a partial hemichrome state | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vergara, A, Franzese, M, Merlino, A, Vitagliano, L, Mazzarella, L. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of ferric hemoglobins from three antarctic fish species of the suborder notothenioidei.
Biophys.J., 93, 2007
|
|
6FPC
 
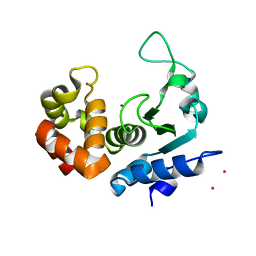 | | Structure of the PRO-PRO endopeptidase (PPEP-2) from Paenibacillus alvei | | Descriptor: | CADMIUM ION, PRO-PRO endopeptidase, SULFATE ION, ... | | Authors: | Weeks, S.D, Klychnikov, O.I, Hensbergen, P.J, Strelkov, S.V. | | Deposit date: | 2018-02-09 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of a new Pro-Pro endopeptidase, PPEP-2, provides mechanistic insights into the differences in substrate specificity within the PPEP family.
J. Biol. Chem., 293, 2018
|
|
4X2E
 
 | | Clostridium difficile wild type Fic protein | | Descriptor: | Fic family protein putative filamentation induced by cAMP protein | | Authors: | Jorgensen, R, Dedic, E. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structure of wild type Clostridium difficile Fic_0569 at 3.1 Angstroms resolution
To Be Published
|
|
4X2C
 
 | | Clostridium difficile Fic protein_0569 mutant S31A, E35A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fic family protein putative filamentation induced by cAMP protein, GLYCEROL, ... | | Authors: | Jorgensen, R, Dedic, E. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Clostridium difficile Fic_0569 S31A, E35A mutant at 1.8 Angstroms resolution
To Be Published
|
|
4X2D
 
 | |
