2IX2
 
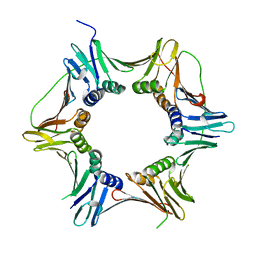 | | Crystal structure of the heterotrimeric PCNA from Sulfolobus solfataricus | | Descriptor: | DNA POLYMERASE SLIDING CLAMP A, DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C | | Authors: | Williams, G.J, Johnson, K, McMahon, S.A, Carter, L, Oke, M, Liu, H, Taylor, G.L, White, M.F, Naismith, J.H. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Heterotrimeric PCNA from Sulfolobus Solfataricus.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1EXP
 
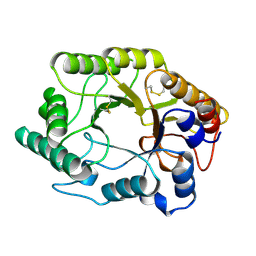 | | BETA-1,4-GLYCANASE CEX-CD | | Descriptor: | BETA-1,4-D-GLYCANASE CEX-CD, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | White, A, Tull, D, Johns, K.L, Withers, S.G, Rose, D.R. | | Deposit date: | 1996-01-11 | | Release date: | 1997-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic observation of a covalent catalytic intermediate in a beta-glycosidase.
Nat.Struct.Biol., 3, 1996
|
|
1FQG
 
 | |
1AP2
 
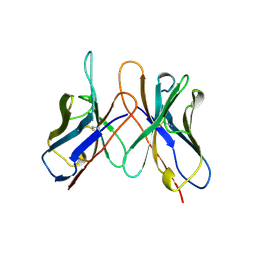 | | SINGLE CHAIN FV OF C219 | | Descriptor: | MONOCLONAL ANTIBODY C219 | | Authors: | Hoedemaeker, P.J, Rose, D.R. | | Deposit date: | 1997-07-23 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A single chain Fv fragment of P-glycoprotein-specific monoclonal antibody C219. Design, expression, and crystal structure at 2.4 A resolution.
J.Biol.Chem., 272, 1997
|
|
1MVU
 
 | |
2AP2
 
 | |
3GMU
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein (BLIP) in Apo Form | | Descriptor: | AMMONIUM ION, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Strynadka, N.C.J, Gretes, M, James, M.N.G. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
1BLN
 
 | | ANTI-P-GLYCOPROTEIN FAB MRK-16 | | Descriptor: | PROTEIN (MONOCLONAL ANTIBODY MRK-16 (HEAVY CHAIN)), PROTEIN (MONOCLONAL ANTIBODY MRK-16 (LIGHT CHAIN)) | | Authors: | Vasudevan, S, Tsuruo, T, Rose, D.R. | | Deposit date: | 1998-07-16 | | Release date: | 1998-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mode of binding of anti-P-glycoprotein antibody MRK-16 to its antigen. A crystallographic and molecular modeling study.
J.Biol.Chem., 273, 1998
|
|
7RWE
 
 | | Crystal structure of CDK2 liganded with compound GPHR787 | | Descriptor: | 5-nitro-2-[(3-phenylpropyl)amino]benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7RWF
 
 | | Crystal structure of CDK2 in complex with TW8672 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
7RXO
 
 | | Crystal structure of CDK2 liganded with compound WN333 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(methoxycarbonyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S4T
 
 | | Crystal structure of CDK2 liganded with compound EF2252 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(6-chloro-1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S84
 
 | | Crystal structure of CDK2 liganded with compound TW8972 | | Descriptor: | 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
7S7A
 
 | | Crystal structure of CDK2 liganded with compound EF3019 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(2H-indazol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S85
 
 | | Crystal structure of CDK2 liganded with compound WN316 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethoxy)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
8D4Z
 
 | | Crystal structure of USP7 in complex with allosteric inhibitor FX1-3763 | | Descriptor: | 1-({(7M)-7-[1-(azetidin-3-yl)-6-chloro-1,2,3,4-tetrahydroquinolin-8-yl]thieno[3,2-b]pyridin-2-yl}methyl)pyrrolidine-2,5-dione, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Bell, J.A. | | Deposit date: | 2022-06-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel USP7 inhibitors demonstrate potent anti-cancer activity in models of AML, synergy with BCL2 inhibition, and a differentiated mechanism of action
To Be Published
|
|
8FOW
 
 | | Ternary complex of CDK2 with small molecule ligands TW8672 and Dinaciclib | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-01-03 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
8FP5
 
 | | CDK2 liganded with ATP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cyclin-dependent kinase 2, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-01-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
8FP0
 
 | |
8PHI
 
 | | Crystal structure of prefusion-stabilized RSV F Variant DS-Cav1 in complex with Lonafarnib | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, CALCIUM ION, ... | | Authors: | Rajak, M, Krey, T. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Drug repurposing screen identifies lonafarnib as respiratory syncytial virus fusion protein inhibitor.
Nat Commun, 15, 2024
|
|
4KNR
 
 | | Hin GlmU bound to WG188 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, N-{4-[(2-benzyl-7-hydroxy-6-methoxyquinazolin-4-yl)amino]phenyl}benzamide, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-10 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
8G21
 
 | |
4KPX
 
 | | Hin GlmU bound to WG766 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, N-{4-[(4-hydroxy-3-nitrobenzoyl)amino]phenyl}pyridine-2-carboxamide, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-14 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
4KPZ
 
 | | Hin GlmU bound to a small molecule fragment | | Descriptor: | 1-(3-nitrophenyl)dihydropyrimidine-2,4(1H,3H)-dione, Bifunctional protein GlmU, MAGNESIUM ION, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-14 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
4KNX
 
 | | Hin GlmU Bound to WG176 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-10 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
