2PDU
 
 | | Human aldose reductase mutant C303D complexed with IDD393. | | Descriptor: | (5-CHLORO-2-{[(3-NITROBENZYL)AMINO]CARBONYL}PHENOXY)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-04-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
6EH3
 
 | |
6EMM
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with Salicylic acid | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, 2-HYDROXYBENZOIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bertoletti, N, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2017-10-02 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | X-ray Crystallographic Fragment screening and Hit Optimization
To Be Published
|
|
6YYZ
 
 | | tRNA-Guanine Transglycosylase (TGT) labeled with 5-fluorotryptophan in co-crystallized complex with 6-amino-2-(methylamino)-4-(2-((2R,3R,4R,5R)-3,4,5-trimethoxytetrahydrofuran-2-yl)ethyl)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-(methylamino)-4-(2-((2R,3R,4R,5R)-3,4,5-trimethoxytetrahydrofuran-2-yl)ethyl)-1H-imidazo[4,5-g]quinazolin-8(7H)-one, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Co-crystallization, 19F NMR and nanoESI-MS reveal dimer disturbing inhibitors and conformational changes at dimer contacts
To Be Published
|
|
6Z0D
 
 | |
4H57
 
 | | Thermolysin inhibition | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Englert, L, Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Dissecting the hydrophobic effect on the molecular level: the role of water, enthalpy, and entropy in ligand binding to thermolysin.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2BBF
 
 | | Crystal structure of tRNA-guanine transglycosylase (TGT) from Zymomonas mobilis in complex with 6-amino-3,7-dihydro-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-3,7-DIHYDRO-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2005-10-17 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding.
J.Mol.Biol., 370, 2007
|
|
6YSJ
 
 | | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-1-(4-bromophenyl)ethanone, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N, Paulus, A. | | Deposit date: | 2020-04-22 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10)
To be published
|
|
5HCO
 
 | | Endothiapepsin in complex with hydrazide | | Descriptor: | 4-(dimethylamino)benzohydrazide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
5ISK
 
 | |
5IS4
 
 | |
5ISJ
 
 | |
1Y5W
 
 | | tRNA-guanine Transglycosylase (TGT) in complex with 6-Amino-4-[2-(4-methylphenyl)ethyl]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-[2-(4-METHYLPHENYL)ETHYL]-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
1Y5V
 
 | | tRNA-Guanine Transglycosylase (TGT) in complex with 6-Amino-4-(2-phenylethyl)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-(2-PHENYLETHYL)-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
1Y5X
 
 | | tRNA-guanine Transglycosylase (TGT) in complex with 6-Amino-4-[2-(4-methoxyphenyl)ethyl]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-[2-(4-METHOXYPHENYL)ETHYL]-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
8R1N
 
 | |
8R18
 
 | | Pim1 in complex with (E)-4-(4-hydroxystyryl)benzoic acid and Pimtide | | Descriptor: | (E)-4-(4-hydroxystyryl)benzoic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | What doesn't fit is made to fit: Pim-1 kinase adapts to the configuration of stilbene-based inhibitors.
Arch Pharm, 357, 2024
|
|
8R10
 
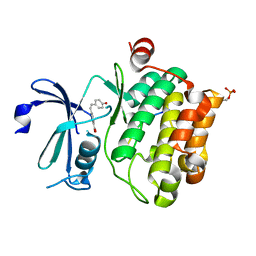 | |
8R1P
 
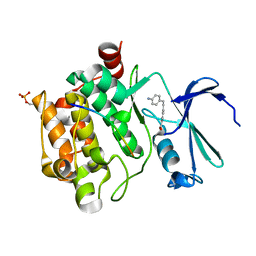 | |
8R1T
 
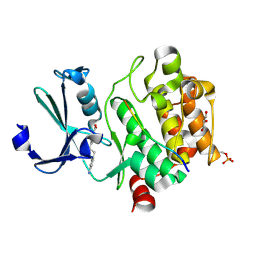 | | Pim1 in complex with 4-(4-aminophenethyl)benzoic acid and Pimtide | | Descriptor: | 4-(4-aminophenethyl)benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | What doesn't fit is made to fit: Pim-1 kinase adapts to the configuration of stilbene-based inhibitors.
Arch Pharm, 357, 2024
|
|
7QB2
 
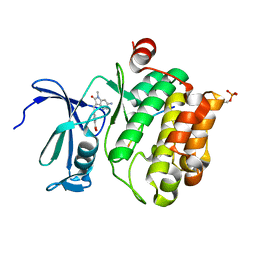 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7QFM
 
 | | Pim1 in complex with (E)-4-((2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
1JGU
 
 | | STRUCTURAL BASIS FOR DISFAVORED ELIMINATION REACTION IN CATALYTIC ANTIBODY 1D4 | | Descriptor: | (2-AMINO-3-PHENYL-BICYCLO[2.2.1]HEPT-2-YL)-PHENYL-METHANONE, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Larsen, N.A, Heine, A, Crane, L, Cravatt, B.F, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2001-06-26 | | Release date: | 2001-12-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for a disfavored elimination reaction in catalytic antibody 1D4.
J.Mol.Biol., 314, 2001
|
|
1JGV
 
 | | STRUCTURAL BASIS FOR DISFAVORED ELIMINATION REACTION IN CATALYTIC ANTIBODY 1D4 | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain | | Authors: | Larsen, N.A, Heine, A, Crane, L, Cravatt, B.F, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2001-06-26 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for a disfavored elimination reaction in catalytic antibody 1D4.
J.Mol.Biol., 314, 2001
|
|
6ZFK
 
 | |
