2MY1
 
 | | Solution structure of Bud31p | | Descriptor: | Pre-mRNA-splicing factor BUD31, ZINC ION | | Authors: | van Roon, A.M, Yang, J, Mathieu, D, Bermel, W, Nagai, K, Neuhaus, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | (113) Cd NMR Experiments Reveal an Unusual Metal Cluster in the Solution Structure of the Yeast Splicing Protein Bud31p.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1XC5
 
 | | Solution Structure of the SMRT Deacetylase Activation Domain | | Descriptor: | Nuclear receptor corepressor 2 | | Authors: | Codina, A, Love, J.D, Li, Y, Lazar, M.A, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2004-09-01 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1XJR
 
 | | The Structure of a Rigorously Conserved RNA Element Within the SARS Virus Genome | | Descriptor: | MAGNESIUM ION, s2m RNA | | Authors: | Robertson, M.P, Igel, H, Baertsch, R, Haussler, D, Ares Jr, M, Scott, W.G. | | Deposit date: | 2004-09-24 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a rigorously conserved RNA element within the SARS virus genome
Plos Biol., 3, 2005
|
|
2JM1
 
 | |
5A3O
 
 | | Crystal structure of the LecB lectin from Pseudomonas aeruginosa in complex with Methyl 6-(cinnamido)-6-deoxy-alpha-D-mannopyranoside at 1.6 ansgtrom | | Descriptor: | CALCIUM ION, CHLORIDE ION, CINNAMIDE, ... | | Authors: | Sommer, R, Hauck, D, Varrot, A, Audfray, A, Imberty, A, Titz, A. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cinnamide Derivatives of D-Mannose as Inhibitors of the Bacterial Virulence Factor Lecb from Pseudomonas Aeruginosa
Chemistryopen, 4, 2015
|
|
1VZS
 
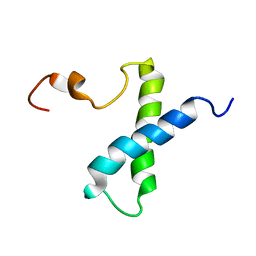 | | Solution structure of subunit F6 from the peripheral stalk region of ATP synthase from bovine heart mitochondria | | Descriptor: | ATP SYNTHASE COUPLING FACTOR 6, MITOCHONDRIAL PRECURSOR | | Authors: | Carbajo, R.J, Silvester, J.A, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Subunit F(6) from the Peripheral Stalk Region of ATP Synthase from Bovine Heart Mitochondria
J.Mol.Biol., 342, 2004
|
|
1URQ
 
 | | Crystal structure of neuronal Q-SNAREs in complex with R-SNARE motif of Tomosyn | | Descriptor: | M-TOMOSYN ISOFORM, SYNAPTOSOMAL-ASSOCIATED PROTEIN 25, SYNTAXIN 1A | | Authors: | Pobbati, A, Razeto, A, Becker, S, Fasshauer, D. | | Deposit date: | 2003-10-31 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibitory Role of Tomosyn in Exocytosis
J.Biol.Chem., 279, 2004
|
|
1UW0
 
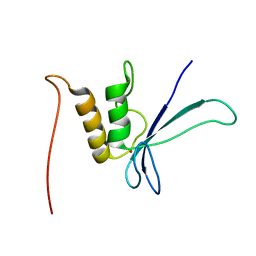 | |
6IAV
 
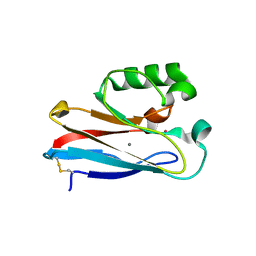 | |
1OAI
 
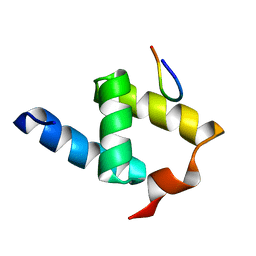 | |
2N1T
 
 | | Dynamic binding mode of a synaptotagmin-1-SNARE complex in solution | | Descriptor: | Synaptosomal-associated protein 25, Synaptotagmin-1, Syntaxin-1A, ... | | Authors: | Brewer, K, Bacaj, T, Cavalli, A, Camilloni, C, Swarbrick, J, Liu, J, Zhou, A, Zhou, P, Barlow, N, Xu, J, Seven, A, Prinslow, E, Voleti, R, Haussinger, D, Bonvin, A, Tomchick, D, Vendruscolo, M, Graham, B, Sudhof, T, Rizo, J. | | Deposit date: | 2015-04-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2K0A
 
 | | 1H, 15N and 13C chemical shift assignments for Rds3 protein | | Descriptor: | Pre-mRNA-splicing factor RDS3, ZINC ION | | Authors: | Loening, N, van Roon, A, Yang, J, Nagai, K, Neuhaus, D. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U2 snRNP protein Rds3p reveals a knotted zinc-finger motif.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3HGJ
 
 | | Old Yellow Enzyme from Thermus scotoductus SA-01 complexed with p-hydroxy-benzaldehyde | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
3HF3
 
 | | Old Yellow Enzyme from Thermus scotoductus SA-01 | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, SULFATE ION | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
1DZ5
 
 | | The NMR structure of the 38KDa U1A protein-PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein | | Descriptor: | PIE, RNA (5'-R(*GP*AP*GP*AP*CP*AP*UP*UP*GP*CP*AP*CP*CP* CP*GP*GP*AP*GP*UP*CP*UP*C)-3'), U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Varani, L, Gunderson, S.I, Mattaj, I.W, Kay, L.E, Neuhaus, D, Varani, G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-03-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the 38kDa U1A Protein-Pie RNA Complex Reveals the Basis of Cooperativity in Regulation of Polyadenylation by Human U1A Protein
Nat.Struct.Biol., 7, 2000
|
|
1FHT
 
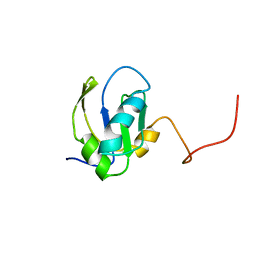 | | RNA-BINDING DOMAIN OF THE U1A SPLICEOSOMAL PROTEIN U1A117, NMR, 43 STRUCTURES | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1996-02-21 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding.
J.Mol.Biol., 257, 1996
|
|
7AAD
 
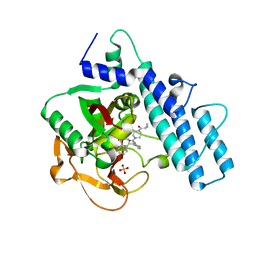 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAA
 
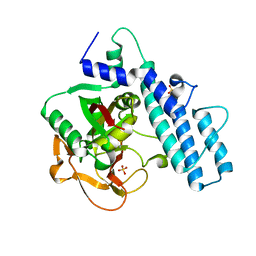 | | Crystal structure of the catalytic domain of human PARP1 (apo) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Underwood, E, Rawlins, P.B, Johannes, J.W, Easton, L.E, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAC
 
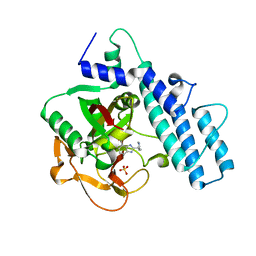 | | Crystal structure of the catalytic domain of human PARP1 in complex with veliparib | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAB
 
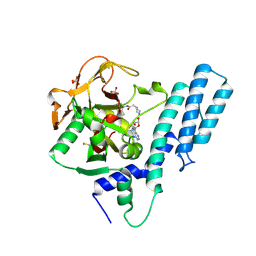 | | Crystal structure of the catalytic domain of human PARP1 in complex with inhibitor EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
6GYI
 
 | |
3MSP
 
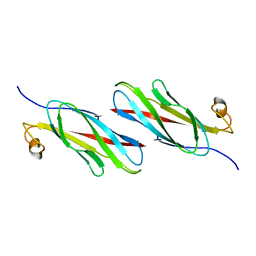 | | MOTILE MAJOR SPERM PROTEIN (MSP) OF ASCARIS SUUM, NMR, 20 STRUCTURES | | Descriptor: | MAJOR SPERM PROTEIN | | Authors: | Haaf, A, Leclaire III, L, Roberts, G, Kent, H.M, Roberts, T.M, Stewart, M, Neuhaus, D. | | Deposit date: | 1998-09-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the motile major sperm protein (MSP) of Ascaris suum - evidence for two manganese binding sites and the possible role of divalent cations in filament formation.
J.Mol.Biol., 284, 1998
|
|
8AUT
 
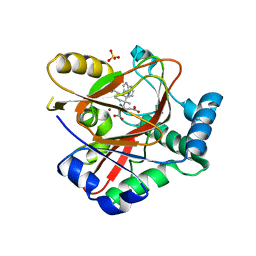 | | WelO5* L221A bound to Zn(II), Cl, 2-oxoglutarate, and 12-epi-hapalindole C | | Descriptor: | 2-OXOGLUTARIC ACID, 3-[(1~{S},2~{R},3~{S},6~{S})-3-ethenyl-2-isocyano-3-methyl-6-prop-1-en-2-yl-cyclohexyl]-1~{H}-indole, CHLORIDE ION, ... | | Authors: | Buller, R, Hueppi, S, Voss, M, Schaub, D. | | Deposit date: | 2022-08-25 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Enzyme engineering enables inversion of substrate stereopreference of the halogenase WelO5*
Chemcatchem, 2022
|
|
2LHN
 
 | | RNA-binding zinc finger protein | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, ZINC ION | | Authors: | Brockmann, C, Neuhaus, D, Stewart, M. | | Deposit date: | 2011-08-12 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Polyadenosine-RNA Binding by Nab2 Zn Fingers and Its Function in mRNA Nuclear Export.
Structure, 20, 2012
|
|
6RH5
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for NECAP1 PHear domain | | Descriptor: | Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
