2ETW
 
 | |
2EVJ
 
 | | Structure of an Ndt80-DNA complex (MSE mutant mA9C) | | Descriptor: | 5'-D(*AP*GP*TP*GP*TP*TP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*AP*AP*CP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
2G9E
 
 | | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TRAM | | Descriptor: | Protein traM | | Authors: | Lu, J, Edwards, R.A, Wong, J.J, Manchak, J, Scott, P.G, Frost, L.S, Glover, J.N. | | Deposit date: | 2006-03-06 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TraM.
Embo J., 25, 2006
|
|
4QPO
 
 | | Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM | | Descriptor: | PHOSPHATE ION, Relaxosome protein TraM | | Authors: | Peng, Y, Lu, J, Wong, J, Edwards, R.A, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Mechanistic Basis of Plasmid-Specific DNA Binding of the F Plasmid Regulatory Protein, TraM.
J.Mol.Biol., 426, 2014
|
|
4QPQ
 
 | | Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM | | Descriptor: | Relaxosome protein TraM, sbmA DNA1, sbmA DNA2 | | Authors: | Peng, Y, Lu, J, Wong, J, Edwards, R.A, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Mechanistic Basis of Plasmid-Specific DNA Binding of the F Plasmid Regulatory Protein, TraM.
J.Mol.Biol., 426, 2014
|
|
4ONL
 
 | | Crystal structure of human Mms2/Ubc13_D81N, R85S, A122V, N123P | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2 | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
4ORH
 
 | | Crystal structure of RNF8 bound to the UBC13/MMS2 heterodimer | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.802 Å) | | Cite: | Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation.
J.Biol.Chem., 287, 2012
|
|
1JNX
 
 | |
1A02
 
 | | STRUCTURE OF THE DNA BINDING DOMAINS OF NFAT, FOS AND JUN BOUND TO DNA | | Descriptor: | AP-1 FRAGMENT FOS, AP-1 FRAGMENT JUN, DNA (5'-D(*DAP*DAP*DCP*DTP*DAP*DTP*DGP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DCP*DC)-3'), ... | | Authors: | Chen, L, Glover, J.N.M, Hogan, P.G, Rao, A, Harrison, S.C. | | Deposit date: | 1997-12-08 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA.
Nature, 392, 1998
|
|
1MNN
 
 | | Structure of the sporulation specific transcription factor Ndt80 bound to DNA | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*TP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*AP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Stuart, D, Tsang, R, Wu, C, Glover, J.N. | | Deposit date: | 2002-09-05 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the sporulation-specific transcription factor Ndt80 bound to DNA
Embo J., 21, 2002
|
|
4KQD
 
 | |
9BIV
 
 | |
1T2V
 
 | | Structural basis of phospho-peptide recognition by the BRCT domain of BRCA1, structure with phosphopeptide | | Descriptor: | BRCTide-7PS, Breast cancer type 1 susceptibility protein | | Authors: | Williams, R.S, Lee, M.S, Hau, D.D, Glover, J.N.M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T2U
 
 | | Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1: structure of BRCA1 missense variant V1809F | | Descriptor: | Breast cancer type 1 susceptibility protein, COBALT (II) ION, SULFATE ION | | Authors: | Williams, R.S, Lee, M.S, Duong, D.D, Glover, J.N.M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Phosphopeptide recognition by the BRCT domain of BRCA1
Nat.Struct.Mol.Biol., 11, 2004
|
|
2R1Z
 
 | |
7RGS
 
 | | The crystal structure of RocC, containing FinO domain, 24-126 | | Descriptor: | Repressor of competence, RNA Chaperone | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
7RGT
 
 | | The crystal structure of RocC, containing FinO domain, 1-126 | | Descriptor: | Repressor of competence, RNA Chaperone, SULFATE ION | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
7RGU
 
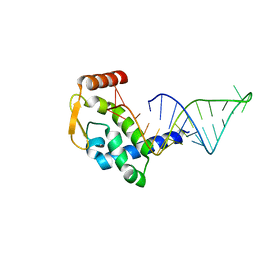 | |
8D34
 
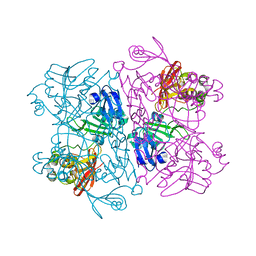 | |
2F9D
 
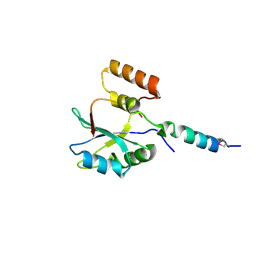 | | 2.5 angstrom resolution structure of the spliceosomal protein p14 bound to region of SF3b155 | | Descriptor: | Pre-mRNA branch site protein p14, Splicing factor 3B subunit 1 | | Authors: | Schellenberg, M.J, Edwards, R.A, Ritchie, D.B, Glover, J.N.M, Macmillan, A.M. | | Deposit date: | 2005-12-05 | | Release date: | 2006-01-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a core spliceosomal protein interface
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1MN4
 
 | | Structure of Ndt80 (Residues 59-340) DNA-binding domain core | | Descriptor: | NDT80 PROTEIN | | Authors: | Lamoureux, J.S, Stuart, D, Tsang, R, Wu, C, Glover, J.N.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the sporulation-specific transcription factor Ndt80 bound to DNA
Embo J., 21, 2002
|
|
1N5O
 
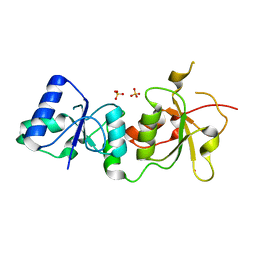 | |
3HUF
 
 | | Structure of the S. pombe Nbs1-Ctp1 complex | | Descriptor: | DNA repair and telomere maintenance protein nbs1, Double-strand break repair protein ctp1, THIOCYANATE ION | | Authors: | Williams, R.S, Guenther, G, Tainer, J.A. | | Deposit date: | 2009-06-13 | | Release date: | 2009-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair.
Cell(Cambridge,Mass.), 139, 2009
|
|
3HUE
 
 | |
3L11
 
 | | Crystal Structure of the Ring Domain of RNF168 | | Descriptor: | E3 ubiquitin-protein ligase RNF168, MALONATE ION, ZINC ION | | Authors: | Neculai, D, Yermekbayeva, L, Crombet, L, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-10 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation.
J.Biol.Chem., 287, 2012
|
|
