7BY7
 
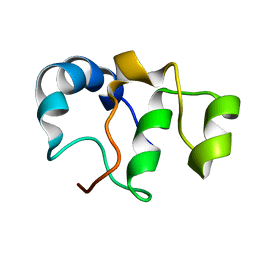 | | Bacteriophage SPO1 protein Gp46 | | Descriptor: | Putative gene 46 protein | | Authors: | Liu, B, Zhang, P. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Bacteriophage protein Gp46 is a cross-species inhibitor of nucleoid-associated HU proteins
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6ZDJ
 
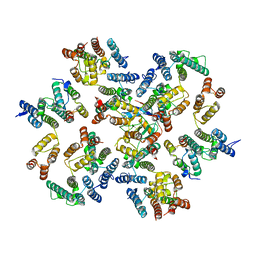 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | Descriptor: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-06-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZET
 
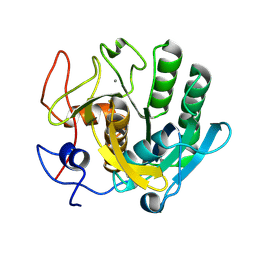 | | Crystal structure of proteinase K nanocrystals by electron diffraction with a 20 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.701 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
6ZEV
 
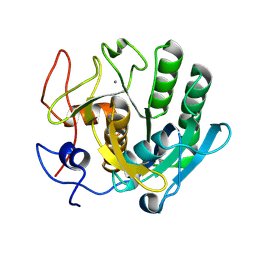 | | Crystal structure of proteinase K lamellae by electron diffraction with a 20 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.4 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
8B12
 
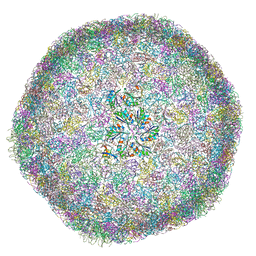 | | cryo-EM structure of carboxysomal mini-shell: icosahedral assembly from CsoS4A/1A and CsoS2 co-expression (T = 9) | | Descriptor: | Carboxysome assembly protein CsoS2B, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Ni, T, Jiang, Q, Liu, L.N, Zhang, P. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.86 Å) | | Cite: | Intrinsically disordered CsoS2 acts as a general molecular thread for alpha-carboxysome shell assembly.
Nat Commun, 14, 2023
|
|
8B0Y
 
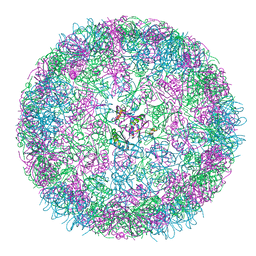 | | cryo-EM structure of carboxysomal mini-shell: icosahedral assembly from CsoS4A/1A co-expression (T = 3) | | Descriptor: | Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Ni, T, Jiang, Q, Liu, L.N, Zhang, P. | | Deposit date: | 2022-09-08 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Intrinsically disordered CsoS2 acts as a general molecular thread for alpha-carboxysome shell assembly.
Nat Commun, 14, 2023
|
|
8B11
 
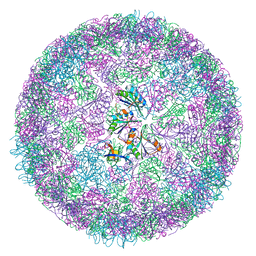 | | cryo-EM structure of carboxysomal mini-shell: icosahedral assembly from CsoS4A/1A and CsoS2 co-expression (T = 4) | | Descriptor: | Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Ni, T, Jiang, Q, Liu, L.N, Zhang, P. | | Deposit date: | 2022-09-08 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Intrinsically disordered CsoS2 acts as a general molecular thread for alpha-carboxysome shell assembly.
Nat Commun, 14, 2023
|
|
6ZEU
 
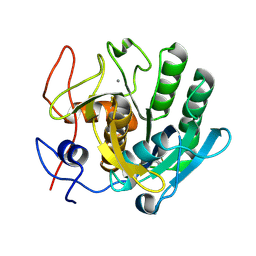 | | Crystal structure of proteinase K lamella by electron diffraction with a 50 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.004 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
6LZ7
 
 | |
6KZ8
 
 | | Crystal structure of plant Phospholipase D alpha complex with phosphatidic acid | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
6KZ9
 
 | | Crystal structure of plant Phospholipase D alpha | | Descriptor: | CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
5XSD
 
 | | XylFII-LytSN complex mutant - D103A | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-13 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XSS
 
 | | XylFII molecule | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, beta-D-xylopyranose | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3H4M
 
 | | AAA ATPase domain of the proteasome- activating nucleotidase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Proteasome-activating nucleotidase | | Authors: | Jeffrey, P, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
1ZUC
 
 | | Progesterone receptor ligand binding domain in complex with the nonsteroidal agonist tanaproget | | Descriptor: | 5-(4,4-DIMETHYL-2-THIOXO-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-6-YL)-1-METHYL-1H-PYRROLE-2-CARBONITRILE, Progesterone receptor, SULFATE ION | | Authors: | Zhang, Z, Olland, A.M, Zhu, Y, Cohen, J, Berrodin, T, Chippari, S, Appavu, C, Li, S, Wilhem, J, Chopra, R, Fensome, A, Zhang, P, Wrobel, J, Unwalla, R.J, Lyttle, C.R, Winneker, R.C. | | Deposit date: | 2005-05-30 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and pharmacological properties of a potent and selective novel nonsteroidal progesterone receptor agonist tanaproget
J.Biol.Chem., 280, 2005
|
|
6LPW
 
 | |
3H43
 
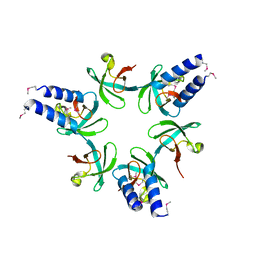 | | N-terminal domain of the proteasome-activating nucleotidase of Methanocaldococcus jannaschii | | Descriptor: | Proteasome-activating nucleotidase | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H4P
 
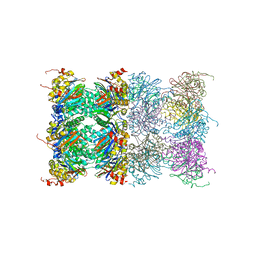 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
4Z7F
 
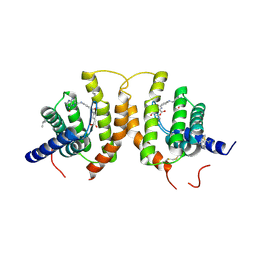 | | Crystal structure of FolT bound with folic acid | | Descriptor: | FOLIC ACID, Folate ECF transporter | | Authors: | Zhao, Q, Wang, C.C, Wang, C.Y, Zhang, P. | | Deposit date: | 2015-04-07 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structures of FolT in substrate-bound and substrate-released conformations reveal a gating mechanism for ECF transporters
Nat Commun, 6, 2015
|
|
6Q0X
 
 | |
4OV9
 
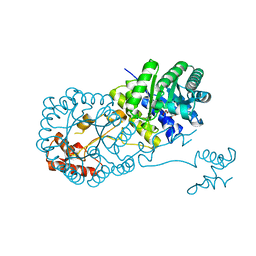 | | Structure of isopropylmalate synthase binding with alpha-isopropylmalate | | Descriptor: | (2S)-2-hydroxy-2-(propan-2-yl)butanedioic acid, ZINC ION, isopropylmalate synthase | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
4OV4
 
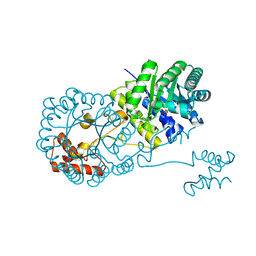 | | Isopropylmalate synthase binding with ketoisovalerate | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, ZINC ION | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
7ASL
 
 | |
7ASH
 
 | |
