4DK5
 
 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine inhibitor | | Descriptor: | 4-(2-[(6-methoxypyridin-3-yl)amino]-5-{[4-(methylsulfonyl)piperazin-1-yl]methyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-based design of a novel series of potent, selective inhibitors of the class I phosphatidylinositol 3-kinases.
J.Med.Chem., 55, 2012
|
|
3S2A
 
 | | Crystal structure of PI3K-gamma in complex with a quinoline inhibitor | | Descriptor: | N-{2-chloro-5-[4-(morpholin-4-yl)quinolin-6-yl]pyridin-3-yl}-4-fluorobenzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phospshoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors: Discovery and Structure-Activity Relationships of a Series of Quinoline and Quinoxaline Derivatives.
J.Med.Chem., 54, 2011
|
|
7CBG
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-N-[(E)-4-[6,7-bis(chloranyl)-4-oxidanylidene-quinazolin-3-yl]but-2-enyl]-2-(methylamino)-3-oxidanyl-butanamide, Threonine--tRNA ligase, ZINC ION | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
7CBI
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)pentyl (2~{S},3~{R})-2-azanyl-3-oxidanyl-butanoate, GLYCEROL, ... | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
6DFI
 
 | |
7CBH
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | Threonine--tRNA ligase, ZINC ION, [(E)-4-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)but-2-enyl] (2S,3R)-2-azanyl-3-oxidanyl-butanoate | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
6DFJ
 
 | |
6KRX
 
 | | Crystal Structure of AtPTP1 at 1.7 angstrom | | Descriptor: | CITRATE ANION, IODIDE ION, Protein-tyrosine-phosphatase PTP1 | | Authors: | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | AtPTP1 positively mediates brassinosteroid signaling from receptor kinases to GSK3-like kinase BIN2
To Be Published
|
|
6KRW
 
 | | Crystal Structure of AtPTP1 at 1.4 angstrom | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, IODIDE ION, ... | | Authors: | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of AtPTP1 at 1.4 Angstroms
To Be Published
|
|
1X79
 
 | | Crystal structure of human GGA1 GAT domain complexed with the GAT-binding domain of Rabaptin5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADP-ribosylation factor binding protein GGA1, Rab GTPase binding effector protein 1, ... | | Authors: | Zhu, G, Zhang, X.C. | | Deposit date: | 2004-08-13 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of human GGA1 GAT domain complexed with the GAT-binding domain of Rabaptin5.
EMBO J., 23, 2004
|
|
1MHQ
 
 | | Crystal Structure Of Human GGA2 VHS Domain | | Descriptor: | ADP-ribosylation factor binding protein GGA2 | | Authors: | Zhu, G, Zhang, X.C. | | Deposit date: | 2002-08-20 | | Release date: | 2003-03-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of GGA2 VHS domain and its implication in plasticity in the ligand binding pocket
FEBS LETT., 537, 2003
|
|
5EHP
 
 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP836 | | Descriptor: | 5-[2,3-bis(chloranyl)phenyl]-2-[(3~{R},5~{S})-3,5-dimethylpiperazin-1-yl]pyrimidin-4-amine, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2015-10-28 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor.
J.Med.Chem., 59, 2016
|
|
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
2M3P
 
 | | DNA containing a cluster of 8-oxo-guanine and abasic site lesion: alpha anomer | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(ORP)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*GP*GP*AP*(8OG)P*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Lourdin, M, Constant, J. | | Deposit date: | 2013-01-25 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
2M40
 
 | | DNA containing a cluster of 8-oxo-guanine and THF lesion | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(3DR)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*GP*GP*AP*(8OG)P*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Constant, J, Lourdin, M. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
2M43
 
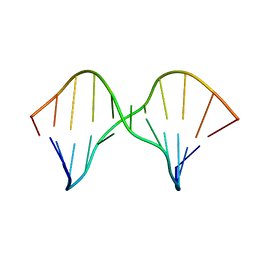 | | DNA containing a cluster of 8-oxo-guanine and abasic site lesion: alpha anomer (AP6, 8OG 14) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(ORP)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*(8OG)P*TP*GP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Constant, J, Lourdin, M. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
2M44
 
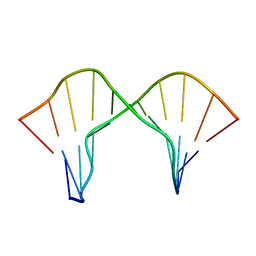 | | DNA containing a cluster of 8-oxo-guanine and abasic site lesion: beta anomer (6AP, 8OG14) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(AAB)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*(8OG)P*TP*GP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Constant, J, Lourdin, M. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
2M3Y
 
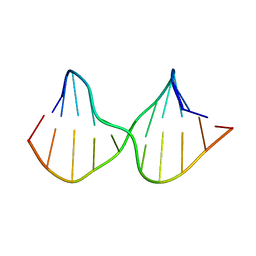 | | DNA containing a cluster of 8-oxo-guanine and abasic site lesion: beta anomer | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(AAB)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*GP*GP*AP*(8OG)P*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Constant, J, Lourdin, M. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
2ER7
 
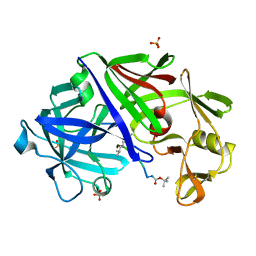 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES.III. THREE-DIMENSIONAL STRUCTURE OF ENDOTHIAPEPSIN COMPLEXED WITH A TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | ENDOTHIAPEPSIN, SULFATE ION, TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN | | Authors: | Veerapandian, B, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-11-12 | | Release date: | 1991-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray analyses of aspartic proteinases. III Three-dimensional structure of endothiapepsin complexed with a transition-state isostere inhibitor of renin at 1.6 A resolution.
J.Mol.Biol., 216, 1990
|
|
6KIH
 
 | | Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, Tll1590 protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Su, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Co-crystal Structure ofThermosynechococcus elongatusSucrose Phosphate Synthase With UDP and Sucrose-6-Phosphate Provides Insight Into Its Mechanism of Action Involving an Oxocarbenium Ion and the Glycosidic Bond.
Front Microbiol, 11, 2020
|
|
3DBN
 
 | | Crystal structure of the Streptoccocus suis serotype2 D-mannonate dehydratase in complex with its substrate | | Descriptor: | D-MANNONIC ACID, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q, Gao, F, Gao, G.F. | | Deposit date: | 2008-06-02 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
4FHK
 
 | | Crystal Structure of PI3K-gamma in Complex with Imidazopyridazine 19e | | Descriptor: | 3-[2-methyl-6-(pyrazin-2-ylamino)pyrimidin-4-yl]-N-(1H-pyrazol-3-yl)imidazo[1,2-b]pyridazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Shaffer, P.L, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-06 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and optimization of potent and selective imidazopyridine and imidazopyridazine mTOR inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6LDQ
 
 | |
1SH6
 
 | |
4HGT
 
 | | Crystal structure of ck1d with compound 13 | | Descriptor: | 2-{2-[(3,4-difluorophenoxy)methyl]-5-methoxypyridin-4-yl}-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform delta | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Potent and Selective CK1 gamma Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
