3TB2
 
 | | 1-Cys peroxidoxin from Plasmodium Yoelli | | Descriptor: | 1,2-ETHANEDIOL, 1-Cys peroxiredoxin, GLYCEROL, ... | | Authors: | Qiu, W, Artz, J.D, Vedadi, M, Sharma, S, Houston, S, Lew, J, Wasney, G, Amani, M, Xu, X, Bray, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Hui, R, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-08-04 | | Release date: | 2011-10-19 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
6H38
 
 | | The crystal structure of human carbonic anhydrase VII in complex with 4-[(4-fluorophenyl)methyl]-1-piperazinyl]benzenesulfonamide. | | Descriptor: | 4-[4-[(4-fluorophenyl)methyl]piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 7, GLYCEROL, ... | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
6H36
 
 | | The crystal structure of human carbonic anhydrase VII in complex with 4-(4-phenylpiperidine-1-carbonyl)benzenesulfonamide. | | Descriptor: | 4-(4-phenylpiperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 7, ZINC ION | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
5VVM
 
 | |
6H3T
 
 | | Schmallenberg Virus Glycoprotein Gc Head Domain in Complex with scFv 1C11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hellert, J, Aebischer, A, Wernike, K, Haouz, A, Brocchi, E, Reiche, S, Guardado-Calvo, P, Beer, M, Rey, F.A. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.836 Å) | | Cite: | Orthobunyavirus spike architecture and recognition by neutralizing antibodies.
Nat Commun, 10, 2019
|
|
6H37
 
 | | The crystal structure of human carbonic anhydrase VII in complex with 4-(4-phenyl)-4-hydroxy-1-piperidine-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-(4-oxidanyl-4-phenyl-piperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 7, ZINC ION | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
8B30
 
 | |
6H3S
 
 | | Schmallenberg Virus Glycoprotein Gc Head/Stalk Domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Envelopment polyprotein, ... | | Authors: | Hellert, J, Aebischer, A, Wernike, K, Haouz, A, Brocchi, E, Reiche, S, Guardado-Calvo, P, Beer, M, Rey, F.A. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | Orthobunyavirus spike architecture and recognition by neutralizing antibodies.
Nat Commun, 10, 2019
|
|
6H3X
 
 | | Oropouche Virus Glycoprotein Gc Head Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Polyprotein, ... | | Authors: | Hellert, J, Aebischer, A, Wernike, K, Haouz, A, Brocchi, E, Reiche, S, Guardado-Calvo, P, Beer, M, Rey, F.A. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Orthobunyavirus spike architecture and recognition by neutralizing antibodies.
Nat Commun, 10, 2019
|
|
5A3P
 
 | | Crystal structure of the catalytic domain of human PLU1 (JARID1B). | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Tallant, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
6H3V
 
 | | Bunyamwera Virus Glycoprotein Gc Head Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Envelopment polyprotein | | Authors: | Hellert, J, Aebischer, A, Wernike, K, Haouz, A, Brocchi, E, Reiche, S, Guardado-Calvo, P, Beer, M, Rey, F.A. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Orthobunyavirus spike architecture and recognition by neutralizing antibodies.
Nat Commun, 10, 2019
|
|
6H3W
 
 | | La Crosse Virus Glycoprotein Gc Head Domain | | Descriptor: | Envelopment polyprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hellert, J, Aebischer, A, Wernike, K, Haouz, A, Brocchi, E, Reiche, S, Guardado-Calvo, P, Beer, M, Rey, F.A. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Orthobunyavirus spike architecture and recognition by neutralizing antibodies.
Nat Commun, 10, 2019
|
|
8ATB
 
 | | Discovery of IRAK4 Inhibitor 16 | | Descriptor: | GLYCEROL, Interleukin-1 receptor-associated kinase 4, ~{N}-[6-ethoxy-2-[2-(4-methylpiperazin-1-yl)-2-oxidanylidene-ethyl]indazol-5-yl]-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-08-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
6UXT
 
 | | Crystal structure of unknown function protein yfdX from Shigella flexneri | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Chang, C, Skarina, T, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-08 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Crystal structure of unknown function protein yfdX from Shigella flexneri
To Be Published
|
|
6G3T
 
 | | X-ray structure of NSD3-PWWP1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-26 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
7TUJ
 
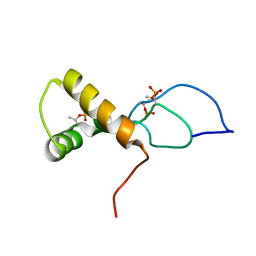 | | NMR solution structure of the phosphorylated MUS81-binding region from human SLX4 | | Descriptor: | Structure-specific endonuclease subunit SLX4 | | Authors: | Payliss, B.J, Reichheld, S.E, Lemak, A, Arrowsmith, C.H, Sharpe, S, Wyatt, H.D.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-09 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease.
Cell Rep, 41, 2022
|
|
1OYZ
 
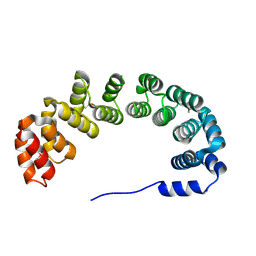 | | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31. | | Descriptor: | Hypothetical protein yibA | | Authors: | Kuzin, A, Semesi, A, Skarina, T, Savchenko, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-12 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31.
To be Published
|
|
3BVN
 
 | | High resolution crystal structure of HLA-B*1402 in complex with the latent membrane protein 2 peptide (LMP2) of Epstein-Barr virus | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B*1402 alpha chain, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2008-01-07 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
1AVH
 
 | | CRYSTAL AND MOLECULAR STRUCTURE OF HUMAN ANNEXIN V AFTER REFINEMENT. IMPLICATIONS FOR STRUCTURE, MEMBRANE BINDING AND ION CHANNEL FORMATION OF THE ANNEXIN FAMILY OF PROTEINS | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Huber, R, Berendes, R, Burger, A, Schneider, M, Karshikov, A, Luecke, H, Roemisch, J, Paques, E. | | Deposit date: | 1991-10-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal and molecular structure of human annexin V after refinement. Implications for structure, membrane binding and ion channel formation of the annexin family of proteins.
J.Mol.Biol., 223, 1992
|
|
6G2F
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 16 | | Descriptor: | 4-[5-(7-fluoranylquinolin-4-yl)-1-methyl-imidazol-4-yl]-3,5-dimethyl-1,2-oxazole, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
1BD6
 
 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
6TAX
 
 | | Mouse RNF213 wild type protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Ahel, J, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.
Elife, 9, 2020
|
|
5CFG
 
 | | C2 crystal form of APE1 with Mg2+ | | Descriptor: | DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural comparison of AP endonucleases from the exonuclease III family reveals new amino acid residues in human AP endonuclease 1 that are involved in incision of damaged DNA.
Biochimie, 128-129, 2016
|
|
457D
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATTCGCG): N6-METHOXYADENOSINE/ THYMIDINE BASE-PAIRS IN B-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(A47)P*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Chatake, T, Ono, A, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 1999-03-06 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on damaged DNAs. II. N(6)-methoxyadenine can present two alternate faces for Watson-Crick base-pairing, leading to pyrimidine transition mutagenesis.
J.Mol.Biol., 294, 1999
|
|
4AII
 
 | | Crystal structure of the rat REM2 GTPase - G domain bound to GDP | | Descriptor: | GTP-BINDING PROTEIN REM 2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Reymond, P, Coquard, A, Chenon, M, Zeghouf, M, El Marjou, A, Thompson, A, Menetrey, J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of the Gdp-Bound G Domain of the Rgk Protein Rem2.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
