6LBH
 
 | |
4UXG
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, R32 native crystal | | Descriptor: | LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
4GLB
 
 | | Structure of p-nitrobenzaldehyde inhibited lipase from Thermomyces lanuginosa at 2.69 A resolution | | Descriptor: | 4-nitrobenzaldehyde, GLYCEROL, Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of p-nitrobenzaldehyde inhibited lipase from Thermomyces lanuginosa at 2.69 A resolution
TO BE PUBLISHED
|
|
4GI1
 
 | | Structure of the complex of three phase partition treated lipase from Thermomyces lanuginosa with 16-hydroxypalmitic acid at 2.4 A resolution | | Descriptor: | 16-hydroxyhexadecanoic acid, GLYCEROL, Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of the complex of three phase partition treated lipase from Thermomyces lanuginosa with 16-hydroxypalmitic acid at 2.4 A resolution
To be published
|
|
4WCC
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation P225G | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-MONOPHOSPHATE, CHLORIDE ION | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WDB
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation R307Q, complexed with 2'-AMP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-MONOPHOSPHATE, CHLORIDE ION | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
6RZU
 
 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
5I05
 
 | |
4GHM
 
 | | Crystal Structure of the H233A mutant of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with preQ0 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Crystal Structure of the H233A mutant of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with preQ0
To be Published
|
|
4UT3
 
 | | X-ray structure of the human PP1 gamma catalytic subunit treated with hydrogen peroxide | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Zeh Silva, M, Kopec, J, Fotinou, D, Steiner, R.A. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Targeted Redox Inhibition of Protein Phosphatase 1 by Nox4 Regulates Eif2Alpha-Mediated Stress Signaling.
Embo J., 35, 2016
|
|
4UT2
 
 | | X-ray structure of the human PP1 gamma catalytic subunit treated with ascorbate | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Kopec, J, Zeh Silva, M, Fotinou, C, Steiner, R.A. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeted Redox Inhibition of Protein Phosphatase 1 by Nox4 Regulates Eif2Alpha-Mediated Stress Signaling.
Embo J., 35, 2016
|
|
4G5Q
 
 | | Structure of LGN GL4/Galphai1 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
4UXF
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 native crystal | | Descriptor: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
2GEJ
 
 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP-Man | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
4GER
 
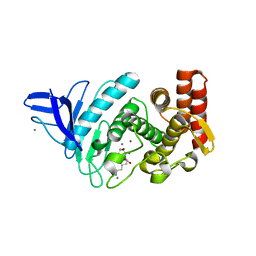 | | Crystal structure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa | | Descriptor: | CALCIUM ION, Gentlyase metalloprotease, LYSINE, ... | | Authors: | Ruf, A, Stihle, M, Benz, J, Schmidt, M, Sobek, H. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6RZT
 
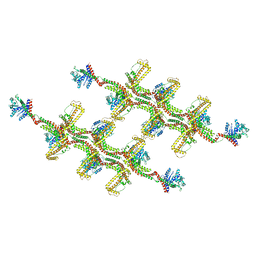 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
4GJ8
 
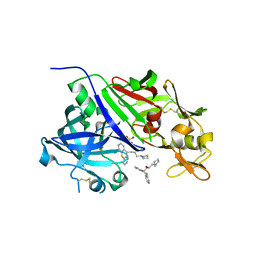 | | Crystal structure of renin in complex with PKF909-724 (compound 3) | | Descriptor: | (2R)-1-(pyrrolidin-1-yl)-3-(9H-thioxanthen-9-yl)propan-2-ol, (2S)-1-(pyrrolidin-1-yl)-3-(9H-thioxanthen-9-yl)propan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2012-08-09 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A novel class of oral direct Renin inhibitors: highly potent 3,5-disubstituted piperidines bearing a tricyclic p3-p1 pharmacophore.
J.Med.Chem., 56, 2013
|
|
2GEK
 
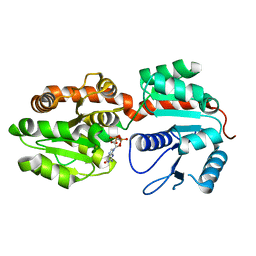 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
6RLC
 
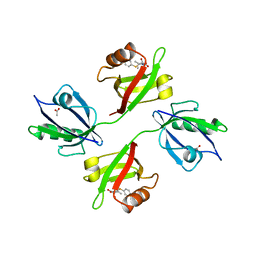 | | Crystal structure of the PDZ tandem of syntenin in complex with fragment F13 | | Descriptor: | (2~{S})-2-[3-(4-chlorophenyl)sulfanylpropanoylamino]-3-methyl-butanoic acid, ACETATE ION, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2019-05-02 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological inhibition of syntenin PDZ2 domain impairs breast cancer cell activities and exosome loadifing with syndecan and EpCAM cargo.
J Extracell Vesicles, 10, 2020
|
|
4GJD
 
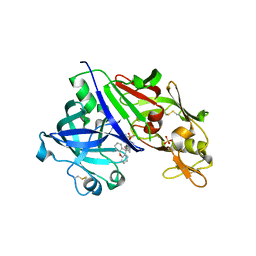 | | Crystal structure of renin in complex with NVP-BGQ311 (compound 12) | | Descriptor: | (3S,5R)-N-{[9-(4-methoxybutyl)-9H-xanthen-9-yl]methyl}-5-{[(4-methylphenyl)sulfonyl]amino}piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2012-08-09 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A novel class of oral direct Renin inhibitors: highly potent 3,5-disubstituted piperidines bearing a tricyclic p3-p1 pharmacophore.
J.Med.Chem., 56, 2013
|
|
7DEV
 
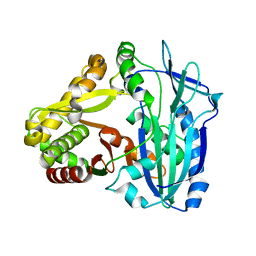 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora | | Descriptor: | Anthocyanin 5-aromatic acyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
7DEX
 
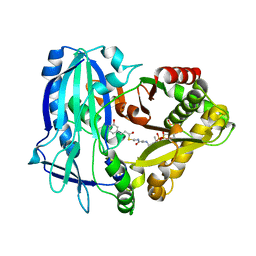 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase H174A mutant with caffeoyl-CoA | | Descriptor: | Anthocyanin 5-aromatic acyltransferase, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (E)-3-[3,4-bis(oxidanyl)phenyl]prop-2-enethioate | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
4WBL
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation F235A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, GLYCEROL | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
7D4V
 
 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase Y91F/C120S variant | | Descriptor: | Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7D5M
 
 | | Crystal structure of inositol dehydrogenase homolog complexed with NAD+ from Azotobacter vinelandii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NAD+ from Azotobacter vinelandii
To Be Published
|
|
