5I9F
 
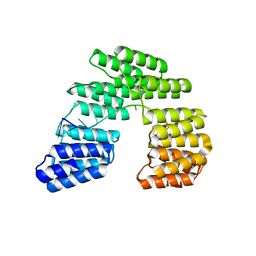 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U10 in complex with its target RNA U10 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U10 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, Y. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
7D12
 
 | | NMR solution structures of CAG RNA-DB213 binding complex | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*CP*AP*GP*C)-3'), SODIUM ION | | Authors: | Chan, H.Y.E, Guo, P. | | Deposit date: | 2020-09-12 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CAG RNAs induce DNA damage and apoptosis by silencing NUDT16 expression in polyglutamine degeneration.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5I9H
 
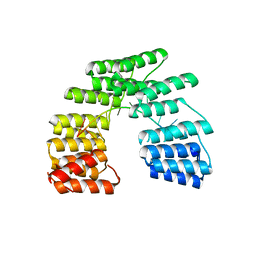 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8G2 in complex with its target RNA U8G2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*GP*GP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8G2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
9IHT
 
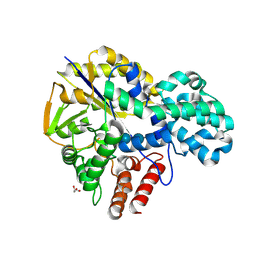 | | Crystal structure of GH57 family amylopullulanase from Aquifex aeolicus in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, Glycoside hydrolase family 57 N-terminal domain-containing protein | | Authors: | Zhu, Z.M, Wang, W.W, Yu, F. | | Deposit date: | 2024-06-18 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | The crystal structure of GH57 family amylopullulanase reveals its dual binding pockets sharing the same catalytic dyad.
Commun Biol, 8, 2025
|
|
9UDE
 
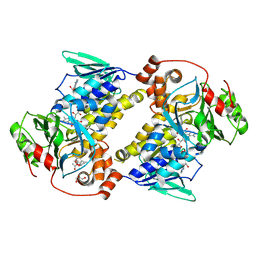 | | Crystal structure of MonCI in complex with diepoxidized farnesyl acetate | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xiao, H.L, Guan, Y.Z, Chen, X. | | Deposit date: | 2025-04-06 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
9UDB
 
 | | Crystal structure of MonCI in complex with farnesyl acetate | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl acetate, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Deng, Y.M, Chen, X. | | Deposit date: | 2025-04-06 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
9UDD
 
 | | Crystal structure of MonCI in complex with monoepoxidized farnesyl acetate | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xiao, H.L, Guan, Y.Z, Chen, X. | | Deposit date: | 2025-04-06 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
6LN1
 
 | | A natural inhibitor of DYRK1A for treatment of diabetes mellitus | | Descriptor: | 1,3,5,8-tetrakis(oxidanyl)xanthen-9-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Li, H, Chen, L.X, Zheng, M.Z, Zhang, Q.Z, Zhang, C.L, Wu, C.R, Yang, K.Y, Song, Z.R, Wang, Q.Q, Li, C, Zhou, Y.R, Chen, J.C. | | Deposit date: | 2019-12-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | A natural DYRK1A inhibitor as a potential stimulator for beta-cell proliferation in diabetes.
Clin Transl Med, 11, 2021
|
|
4RB2
 
 | |
3EIJ
 
 | | Crystal structure of Pdcd4 | | Descriptor: | Programmed cell death protein 4 | | Authors: | Loh, P.G. | | Deposit date: | 2008-09-16 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
3EIQ
 
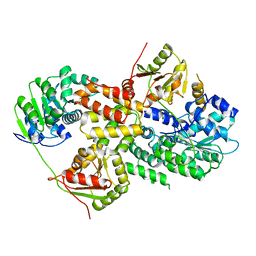 | | Crystal structure of Pdcd4-eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I, Programmed cell death protein 4 | | Authors: | Loh, P.G, Cheng, Z, Song, H. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
4RAY
 
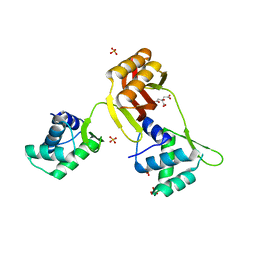 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Apo-Fur | | Descriptor: | CITRATE ANION, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), SULFATE ION | | Authors: | Deng, Z, Liu, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4RB3
 
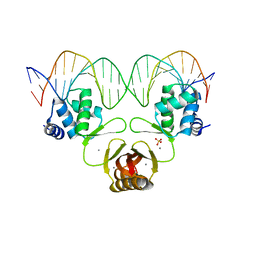 | |
4RB0
 
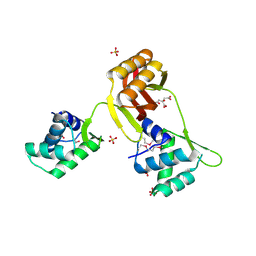 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 SeMet-Apo-Fur | | Descriptor: | CITRATE ANION, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), SULFATE ION | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4RB1
 
 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Fur-Mn2+-E. coli Fur box | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*AP*TP*GP*AP*TP*AP*AP*TP*CP*AP*TP*TP*AP*TP*CP*CP*GP*C)-3'), DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), MANGANESE (II) ION | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6L1H
 
 | |
5XGR
 
 | | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Xue, H, Qi, J, Song, H, Qihui, W, Shi, Y, Gao, G.F. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein
Virology, 507, 2017
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7F9I
 
 | | The apo-form structure of EnrR | | Descriptor: | EnrR repressor | | Authors: | Gan, J.H, Wang, Q.Y. | | Deposit date: | 2021-07-04 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
7F9H
 
 | | complex structure of EnrR-DNA | | Descriptor: | EnrR repressor, target DNA | | Authors: | Gan, J.H, Wang, Q.Y. | | Deposit date: | 2021-07-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
6L1G
 
 | |
9II5
 
 | | Crystal structure of human TRIM21 PRYSPRY in complex with compound 1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, ~{N}-[(1-fluoranylcyclohexyl)methyl]-~{N}-methyl-4-(2-methylsulfanylphenyl)-2-methylsulfonyl-benzamide | | Authors: | Zhang, L.Y. | | Deposit date: | 2024-06-19 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Chemically Induced Nuclear Pore Complex Protein Degradation via TRIM21.
Acs Chem.Biol., 20, 2025
|
|
5OVQ
 
 | |
3NGN
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CCR4-NOT transcription complex subunit 6-like | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
