3X2I
 
 | | X-ray structure of PcCel45A N92D apo form at 298K. | | Descriptor: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2H
 
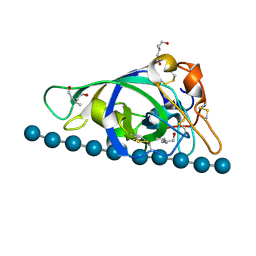 | | X-ray structure of PcCel45A N92D with cellopentaose at 95K. | | Descriptor: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2J
 
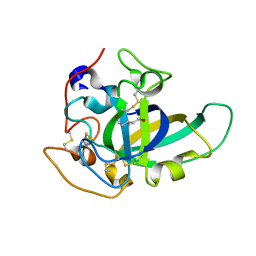 | | X-ray structure of PcCel45A D114N apo form at 95K. | | Descriptor: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2G
 
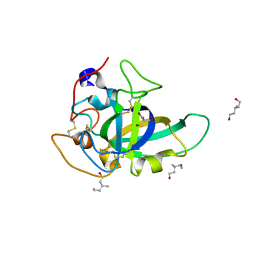 | | X-ray structure of PcCel45A N92D apo form at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2L
 
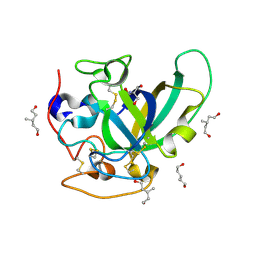 | | X-ray structure of PcCel45A apo form at 95K. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Ohta, K, Tanaka, H, Inaka, K, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2N
 
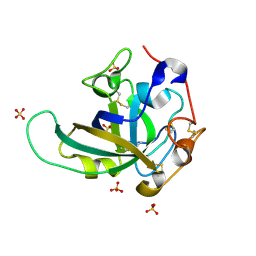 | | Proton relay pathway in inverting cellulase | | Descriptor: | Endoglucanase V-like protein, SULFATE ION | | Authors: | Nakamura, A, Ishida, T, Fushinobu, S, Igarashi, K, Samejima, M. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
6IS6
 
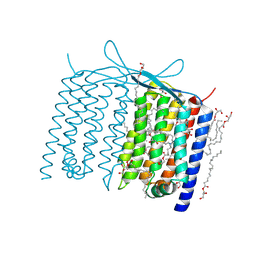 | | Crystal structure of Thermoplasmatales archaeon heliorhodopsin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, RETINAL, heliorhodopsin | | Authors: | Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of heliorhodopsin.
Nature, 574, 2019
|
|
8W6X
 
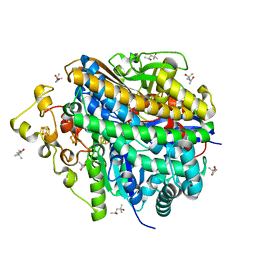 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
6IS0
 
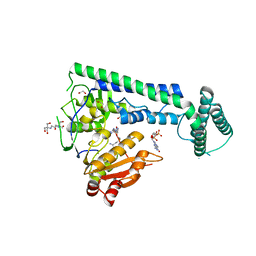 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRW
 
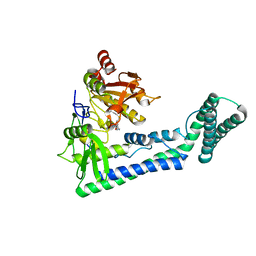 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRX
 
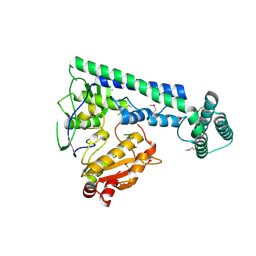 | |
6IRY
 
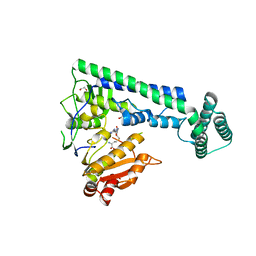 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
8XHR
 
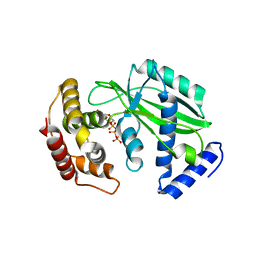 | |
4X8Y
 
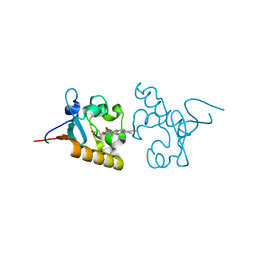 | | Crystal structure of human PGRMC1 cytochrome b5-like domain | | Descriptor: | Membrane-associated progesterone receptor component 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakane, T, Yamamoto, T, Shimamura, T, Kobayashi, T, Kabe, Y, Suematsu, M. | | Deposit date: | 2014-12-11 | | Release date: | 2016-03-23 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance
Nat Commun, 7, 2016
|
|
7L7V
 
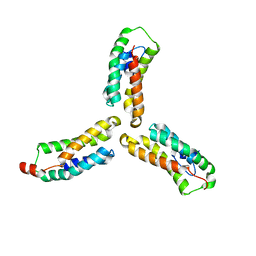 | | Crystal structure of Arabidopsis NRG1.1 CC-R domain K94E/K96E/R99E/K100E/R103E/K106E/K110E mutant | | Descriptor: | Probable disease resistance protein At5g66900 | | Authors: | Walton, W.G, Wan, L, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-30 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plant "helper" immune receptors are Ca 2+ -permeable nonselective cation channels.
Science, 373, 2021
|
|
7L7W
 
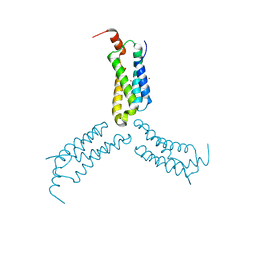 | | Crystal structure of Arabidopsis NRG1.1 CC-R domain K94E/K96E mutant | | Descriptor: | NICKEL (II) ION, Probable disease resistance protein At5g66900 | | Authors: | Walton, W.G, Wan, L, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Plant "helper" immune receptors are Ca 2+ -permeable nonselective cation channels.
Science, 373, 2021
|
|
6EEM
 
 | | Crystal structure of Papaver somniferum tyrosine decarboxylase in complex with L-tyrosine | | Descriptor: | N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-tyrosine, SULFATE ION, TYROSINE, ... | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61000657 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEQ
 
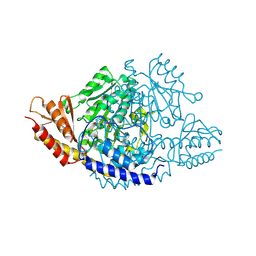 | | Crystal structure of Rhodiola rosea 4-hydroxyphenylacetaldehyde synthase | | Descriptor: | 4-hydroxyphenylacetaldehyde synthase | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.600086 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEI
 
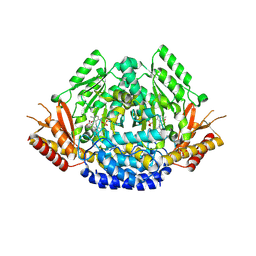 | | Crystal structure of Arabidopsis thaliana phenylacetaldehyde synthase in complex with L-phenylalanine | | Descriptor: | PHENYLALANINE, SULFATE ION, Tyrosine decarboxylase 1 | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (1.99001348 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEW
 
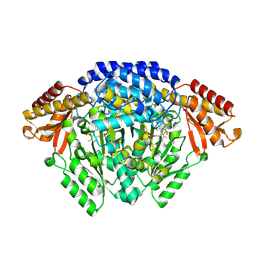 | | Crystal structure of Catharanthus roseus tryptophan decarboxylase in complex with L-tryptophan | | Descriptor: | Aromatic-L-amino-acid decarboxylase, CALCIUM ION, TRYPTOPHAN | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.05002069 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YPE
 
 | |
8U08
 
 | |
8U03
 
 | |
8TZN
 
 | |
8TZW
 
 | |
