7DUZ
 
 | |
7DU1
 
 | |
7DUE
 
 | |
7DUB
 
 | |
7SJO
 
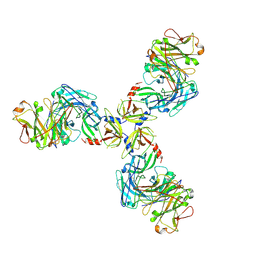 | | HtrA1S328A:Fab15H6.v4 complex | | Descriptor: | Fab15H6.v4 Heavy Chain, Fab15H6.v4 Light Chain, Serine protease HTRA1 | | Authors: | Gerhardy, S, Green, E, Estevez, A, Arthur, C.P, Ultsch, M, Rohou, A, Kirchhofer, D. | | Deposit date: | 2021-10-18 | | Release date: | 2022-09-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric inhibition of HTRA1 activity by a conformational lock mechanism to treat age-related macular degeneration.
Nat Commun, 13, 2022
|
|
8HUX
 
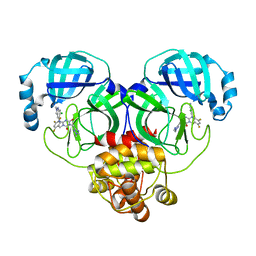 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
9J9J
 
 | | Nitrophenol monooxygenase RsPNPA from Rhodococcus sp. 21391 | | Descriptor: | 4-nitrophenol 4-monooxygenase/4-nitrocatechol 2-monooxygenase, oxygenase component | | Authors: | Yang, J, Li, R, Lin, S, Long, L. | | Deposit date: | 2024-08-22 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biodegradation of p-nitrophenol by Rhodococcus sp. 21391 unveils a two-component p-nitrophenol monooxygenase with broad substrate specificity.
Microb Cell Fact, 24, 2025
|
|
4OE1
 
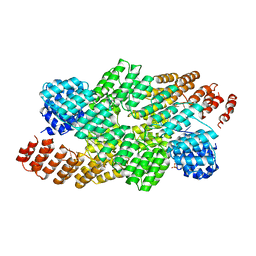 | | Crystal structure of the pentatricopeptide repeat protein PPR10 (C256S/C430S/C449S) in complex with an 18-nt PSAJ rna element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Li, Q, Yan, C, Wu, J, Yin, P, Yan, N. | | Deposit date: | 2014-01-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Examination of the dimerization states of the single-stranded RNA recognition protein pentatricopeptide repeat 10 (PPR10).
J.Biol.Chem., 289, 2014
|
|
7V7M
 
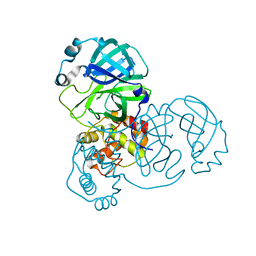 | | crystal structure of SARS-CoV-2 3CL protease | | Descriptor: | 3C-like proteinase | | Authors: | Yi, Y, Zhang, M, Ye, M. | | Deposit date: | 2021-08-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Schaftoside inhibits 3CL pro and PL pro of SARS-CoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19.
Acta Pharm Sin B, 12, 2022
|
|
3HIA
 
 | | Crystal structure of the choline binding domain of Spr1274 in Streptococcus pneumoniae | | Descriptor: | Choline binding protein, PHOSPHOCHOLINE, SULFATE ION | | Authors: | Zhang, Z.-Y, Li, W.-Z, Jiang, Y.-L, Bao, R, Zhou, C.-Z, Chen, Y.-X. | | Deposit date: | 2009-05-19 | | Release date: | 2009-08-04 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of the choline-binding domain of Spr1274 in Streptococcus pneumoniae.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6Y9X
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,7) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9Z
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,9) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YJ5
 
 | |
7ESX
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wPip | | Descriptor: | Bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7X83
 
 | | Cryo-EM structure of the TMEM106B fibril from normal elder | | Descriptor: | Transmembrane protein 106B | | Authors: | Xia, W.C, Zhao, Q.Y, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
7X84
 
 | | Cryo-EM structure of the TMEM106B fibril from Parkinson's disease dementia | | Descriptor: | Transmembrane protein 106B | | Authors: | Zhao, Q.Y, Xia, W.C, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
7PNQ
 
 | |
7PNM
 
 | |
8JPZ
 
 | | The thermostability mutant Gox_M8 from Aspergillus niger | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tu, T, Yan, Y.R. | | Deposit date: | 2023-06-13 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Revealing the intricate mechanism governing the pH-dependent activity of a quintessential representative of flavoproteins, glucose oxidase
Fundam Res, 2024
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7YJO
 
 | | Cryo-EM structure of the monomeric atSPT-ORM1 (LCB2a-deltaN5) complex | | Descriptor: | Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJM
 
 | | Cryo-EM structure of the monomeric atSPT-ORM1 complex | | Descriptor: | Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJN
 
 | | Cryo-EM structure of the monomeric atSPT-ORM1 (ORM1-N17A) complex | | Descriptor: | Long chain base biosynthesis protein 1, Long chain base biosynthesis protein 2a, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJK
 
 | | Cryo-EM structure of the dimeric atSPT-ORM1 complex | | Descriptor: | Long chain base biosynthesis protein 1, Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
6ZDJ
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | Descriptor: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-06-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
