8X7I
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by intein-based E2-Ub-NCP conjugation strategy | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7K
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by activity-based chemical trapping strategy (adjacent H2AK13/15 dual-monoubiquitination) | | Descriptor: | DNA (143-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7J
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub/nucleosomes complex determined by activity-based chemical trapping strategy | | Descriptor: | DNA (144-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8YW6
 
 | | Cryo-EM structure of apo human mitochondrial pyruvate carrier in the IMS-open conformation at pH 8.0 | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-03-29 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
8YW9
 
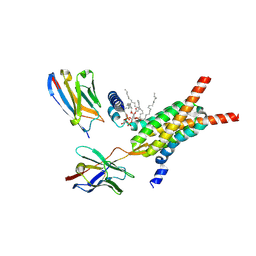 | | Cryo-EM structure of human mitochondrial pyruvate carrier in the matrix-facing conformation at pH 6.8 | | Descriptor: | CARDIOLIPIN, MPC specific nanobody 1, MPC specific nanobody 2, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-03-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
8YW8
 
 | | Cryo-EM structure of human mitochondrial pyruvate carrier in complex with the inhibitor UK5099 | | Descriptor: | (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-03-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
8XMC
 
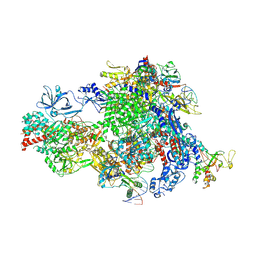 | | Post-translocated Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
8XMB
 
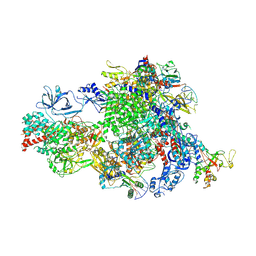 | | NTP-bound Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
8XMD
 
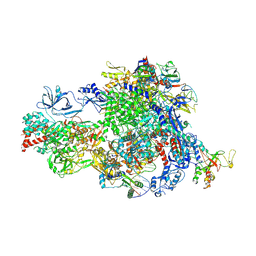 | | Pre-translocated Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
8XME
 
 | | Backtracked Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
8YSF
 
 | | MERS-CoV RBD in complex with nanobody Nb9 | | Descriptor: | Nb9, Spike glycoprotein | | Authors: | Wang, Y.X, Ma, S. | | Deposit date: | 2024-03-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure defining of ultrapotent neutralizing nanobodies against MERS-CoV with novel epitopes on receptor binding domain.
Plos Pathog., 20, 2024
|
|
8YXI
 
 | | Crystal structure of SFTSV Gn in complex with a neutralizing antibody 40C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, P, Guo, Y, Zhang, N. | | Deposit date: | 2024-04-02 | | Release date: | 2024-09-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism and structure-guided humanization of a broadly neutralizing antibody against SFTSV.
Plos Pathog., 20, 2024
|
|
8YSH
 
 | | MERS-CoV RBD in complex with nanobody Nb14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb14, Spike glycoprotein, ... | | Authors: | Wang, Y.X, Ma, S. | | Deposit date: | 2024-03-23 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure defining of ultrapotent neutralizing nanobodies against MERS-CoV with novel epitopes on receptor binding domain.
Plos Pathog., 20, 2024
|
|
9B19
 
 | | EV-D68 in complex with inhibitor Jun11-54-1 | | Descriptor: | Capsid protein VP1, Capsid protein VP4, N-{(4M)-4-[5-(aminomethyl)thiophen-2-yl]quinolin-8-yl}-4-[(propan-2-yl)oxy]benzamide, ... | | Authors: | Klose, T, Kuhn, R.J, Jun, W. | | Deposit date: | 2024-03-13 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Design of enterovirus D68 capsid inhibitors with in vivo antiviral efficacy
To Be Published
|
|
9B1B
 
 | | EV-D68 in complex with inhibitor Jun11-78-7 | | Descriptor: | 4-[(propan-2-yl)oxy]-N-{(4M)-4-[1-(2,2,2-trifluoroethyl)-1H-pyrazol-4-yl]quinolin-8-yl}benzamide, Capsid protein VP1, Capsid protein VP4, ... | | Authors: | Klose, T, Kuhn, R.J, Jun, W. | | Deposit date: | 2024-03-13 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Design of enterovirus D68 capsid inhibitors with in vivo antiviral efficacy
To Be Published
|
|
9B18
 
 | | EV-D68 in complex with inhibitor Jun11-53-7 | | Descriptor: | (4M)-4-[1-(2-methoxyethyl)-1H-pyrazol-4-yl]-N-({4-[(propan-2-yl)oxy]phenyl}methyl)quinolin-8-amine, Capsid protein VP1, Capsid protein VP4, ... | | Authors: | Klose, T, Kuhn, R.J, Jun, W. | | Deposit date: | 2024-03-13 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Design of enterovirus D68 capsid inhibitors with in vivo antiviral efficacy
To Be Published
|
|
9B1A
 
 | | EV-D68 in complex with inhibitor Jun11-69-5 | | Descriptor: | (5M)-5-{8-[({4-[(propan-2-yl)oxy]phenyl}methyl)amino]quinolin-4-yl}pyridine-2-carbonitrile, Capsid protein VP1, Capsid protein VP4, ... | | Authors: | Klose, T, Kuhn, R.J, Jun, W. | | Deposit date: | 2024-03-13 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Design of enterovirus D68 capsid inhibitors with in vivo antiviral efficacy
To Be Published
|
|
7C17
 
 | |
7VF6
 
 | | The crystal structure of PurZ0 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Tong, Y, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Alternative Z-genome biosynthesis pathway shows evolutionary progression from Archaea to phage.
Nat Microbiol, 8, 2023
|
|
5GIY
 
 | | HSA-Palmitic acid-[RuCl5(ind)]2- | | Descriptor: | PALMITIC ACID, Serum albumin, pentakis(chloranyl)-(1~{H}-indazol-2-ium-2-yl)ruthenium(1-) | | Authors: | Yang, F, Wang, T. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Structure of HSA-Palmitic acid-[RuCl5(ind)]2-
To Be Published
|
|
7VF9
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7F3N
 
 | | Structure of PopP2 in apo form | | Descriptor: | Type III effector protein popp2 | | Authors: | Xia, Y, Zhang, Z.M. | | Deposit date: | 2021-06-16 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.351856 Å) | | Cite: | Secondary-structure switch regulates the substrate binding of a YopJ family acetyltransferase.
Nat Commun, 12, 2021
|
|
4JPW
 
 | | Crystal structure of broadly and potently neutralizing antibody 12a21 in complex with hiv-1 strain 93th057 gp120 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEAVY CHAIN OF ANTIBODY 12A21, ... | | Authors: | Acharya, P, Luongo, T, Zhou, T, Kwong, P.D. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization.
Cell(Cambridge,Mass.), 153, 2013
|
|
5T8B
 
 | |
5T8C
 
 | |
