6EHR
 
 | |
8T4C
 
 | |
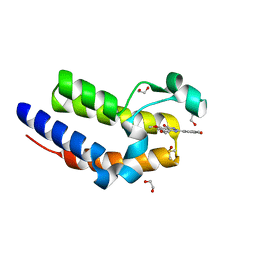
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
4MR5
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with a quinazolinone ligand (RVX-OH) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 2, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8JKK
 
 | |
5ONI
 
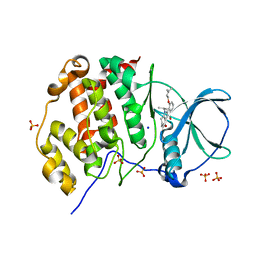 | | LOW-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,4-BUTANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
8K79
 
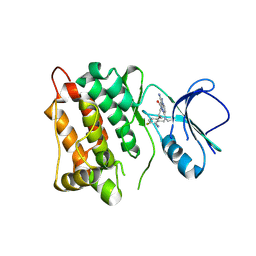 | |
8K78
 
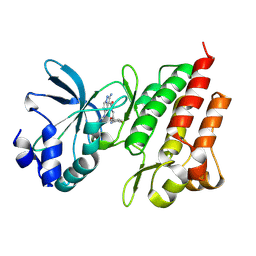 | |
4MR3
 
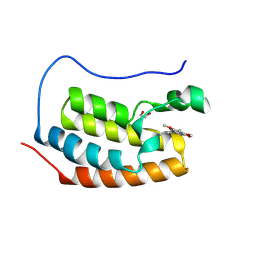 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolinone ligand (RVX-OH) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MR6
 
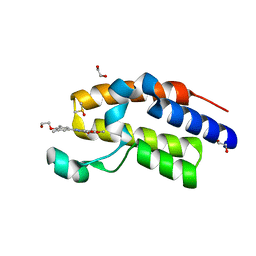 | | Crystal Structure of the second bromodomain of human BRD2 in complex with a quinazolinone ligand (RVX-208) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 2, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MR4
 
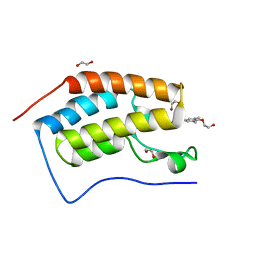 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolinone ligand (RVX-208) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8JFG
 
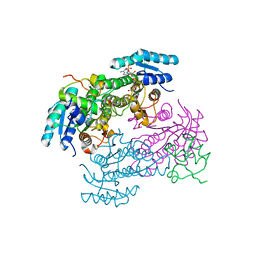 | |
8JFN
 
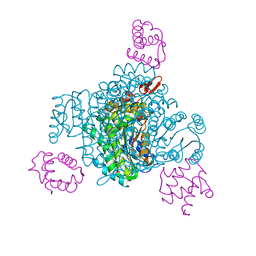 | |
8JFM
 
 | |
8JFJ
 
 | | Crystal structure of enoyl-ACP reductase FabI from Helicobacter pylori | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Song, W.Y, Zhang, L. | | Deposit date: | 2023-05-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Molecular Basis of Catalysis by SDR Family Members Ketoacyl-ACP Reductase FabG and Enoyl-ACP Reductase FabI in Type-II Fatty Acid Biosynthesis.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JFA
 
 | |
8JFH
 
 | |
8JF9
 
 | |
8JFI
 
 | |
8JNK
 
 | |
5GY1
 
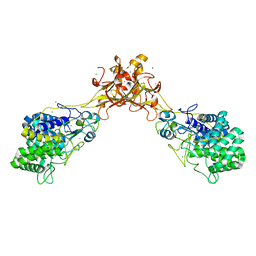 | | Crystal structure of endoglucanase CelQ from Clostridium thermocellum complexed with cellotriose | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structures of the C-Terminally Truncated Endoglucanase Cel9Q from Clostridium thermocellum Complexed with Cellodextrins and Tris.
Chembiochem, 20, 2019
|
|
8JNR
 
 | |
5GXX
 
 | | Crystal structure of endoglucanase CelQ from Clostridium thermocellum complexed with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the C-Terminally Truncated Endoglucanase Cel9Q from Clostridium thermocellum Complexed with Cellodextrins and Tris.
Chembiochem, 20, 2019
|
|
5GXZ
 
 | | Crystal structure of endoglucanase CelQ from Clostridium thermocellum complexed with cellobiose and cellotriose | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of the C-Terminally Truncated Endoglucanase Cel9Q from Clostridium thermocellum Complexed with Cellodextrins and Tris.
Chembiochem, 20, 2019
|
|
8K7I
 
 | | Crystal structure of human lysosomal alpha-galactosidase A in complex with (2R,3S,4R,5R)-2-(hydroxymethyl)-5-((methylamino)methyl)pyrrolidine-3,4-diol | | Descriptor: | (2~{R},3~{S},4~{R},5~{R})-2-(hydroxymethyl)-5-(methylaminomethyl)pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Y, Huang, K.F, Ko, T.P, Cheng, W.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Mechanistic Insights into Dibasic Iminosugars as pH-Selective Pharmacological Chaperones to Stabilize Human alpha-Galactosidase.
Jacs Au, 4, 2024
|
|
