2A7U
 
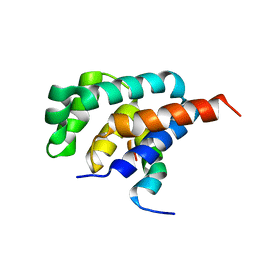 | | NMR solution structure of the E.coli F-ATPase delta subunit N-terminal domain in complex with alpha subunit N-terminal 22 residues | | Descriptor: | ATP synthase alpha chain, ATP synthase delta chain | | Authors: | Wilkens, S, Borchardt, D, Weber, J, Senior, A.E. | | Deposit date: | 2005-07-06 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Interaction of the delta and alpha Subunits of the Escherichia coli F(1)F(0)-ATP Synthase by NMR Spectroscopy
Biochemistry, 44, 2005
|
|
8TAR
 
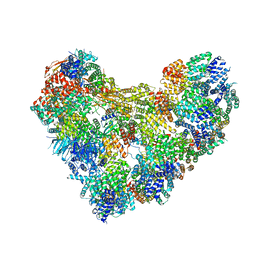 | | APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
8TUP
 
 | | Cryo-EM structure of the human MRS2 magnesium channel under Mg2+-free condition | | Descriptor: | MAGNESIUM ION, Magnesium transporter MRS2 homolog, mitochondrial | | Authors: | Lai, L.T.F, Balaraman, J, Zhou, F, Matthies, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human magnesium channel MRS2 reveal gating and regulatory mechanisms.
Nat Commun, 14, 2023
|
|
8T6V
 
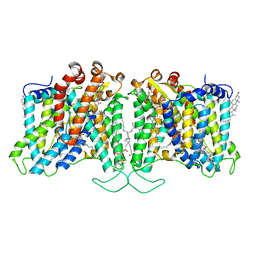 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8TUL
 
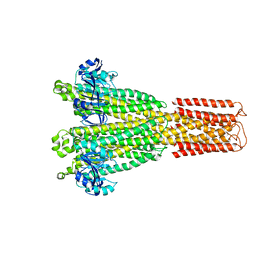 | | Cryo-EM structure of the human MRS2 magnesium channel under Mg2+ condition | | Descriptor: | MAGNESIUM ION, Magnesium transporter MRS2 homolog, mitochondrial | | Authors: | Lai, L.T.F, Balaraman, J, Zhou, F, Matthies, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human magnesium channel MRS2 reveal gating and regulatory mechanisms.
Nat Commun, 14, 2023
|
|
8TAU
 
 | | APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
2Z60
 
 | | Crystal Structure of the T315I Mutant of Abl kinase bound with PPY-A | | Descriptor: | 5-[3-(2-METHOXYPHENYL)-1H-PYRROLO[2,3-B]PYRIDIN-5-YL]-N,N-DIMETHYLPYRIDINE-3-CARBOXAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Dalgarno, D, Zhu, X. | | Deposit date: | 2007-07-21 | | Release date: | 2007-09-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the T315I Mutant of Abl Kinase
Chem.Biol.Drug Des., 70, 2007
|
|
2CSA
 
 | | Structure of the M3 Muscarinic Acetylcholine Receptor Basolateral Sorting Signal | | Descriptor: | Muscarinic acetylcholine receptor M3 | | Authors: | Iverson, H.A, Fox, D, Nadler, L.S, Klevit, R.E, Nathanson, N.M. | | Deposit date: | 2005-05-21 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Identification and structural determination of the M3 muscarinic acetylcholine receptor basolateral sorting signal.
J.Biol.Chem., 280, 2005
|
|
8T6U
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Dipyridamole | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2CTI
 
 | |
2Z2F
 
 | | X-ray Crystal Structure of Bovine Stomach Lysozyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme C-2, SODIUM ION | | Authors: | Akieda, D, Nonaka, Y, Watanabe, N, Tanaka, I, Kamiya, M, Aizawa, T, Nitta, K, Demura, M, Kawano, K. | | Deposit date: | 2007-05-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stability of Bovine Stomach Lysozyme in Acidic Condition
To be Published
|
|
2Z31
 
 | | Crystal structure of immune receptor complex | | Descriptor: | H-2 class II histocompatibility antigen, A-U alpha chain, A-U beta chain precursor, ... | | Authors: | Feng, D, Bond, C.J, Ely, L.K, Garcia, K.C. | | Deposit date: | 2007-05-30 | | Release date: | 2007-10-09 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'.
Nat.Immunol., 8, 2007
|
|
2Z67
 
 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) selenium transferase (SepSecS) | | Descriptor: | O-phosphoseryl-tRNA(Sec) selenium transferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Araiso, Y, Ishitani, R, Pailouer, S, Oshikane, H, Domae, N, Soll, D, Nureki, O. | | Deposit date: | 2007-07-23 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation.
Nucleic Acids Res., 36, 2008
|
|
4CFE
 
 | | Structure of full length human AMPK in complex with a small molecule activator, a benzimidazole derivative (991) | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
2D07
 
 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
2Z2E
 
 | | Crystal Structure of Canine Milk Lysozyme Stabilized against Non-enzymatic Deamidation | | Descriptor: | Lysozyme C, milk isozyme, SULFATE ION | | Authors: | Nonaka, Y, Akieda, D, Watanabe, N, Tanaka, I, Kamiya, M, Aizawa, T, Nitta, K, Demura, M, Kawano, K. | | Deposit date: | 2007-05-21 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Spontaneous asparaginyl deamidation of canine milk lysozyme under mild conditions
Proteins, 72, 2008
|
|
2D0V
 
 | | Crystal structure of methanol dehydrogenase from Hyphomicrobium denitrificans | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, methanol dehydrogenase large subunit, ... | | Authors: | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2005-08-09 | | Release date: | 2006-08-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
2ZGY
 
 | | PARM with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid segregation protein parM | | Authors: | Popp, D, Narita, A, Oda, T, Fujisawa, T, Matsuo, H, Nitanai, Y, Iwasa, M, Maeda, K, Onishi, H, Maeda, Y. | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability
Embo J., 27, 2008
|
|
4BQG
 
 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-(3,4-dichloro-phenoxy)-benzene-1,3-diol, HSP90AA1 PROTEIN | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
2CVI
 
 | | Crystal structure of hypothetical protein PHS023 from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 75aa long hypothetical regulatory protein AsnC, POTASSIUM ION | | Authors: | Kitago, Y, Sakai, N, Matsumoto, D, Yao, M, Aizawa, T, Demura, M, Kawano, K, Nitta, K, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-06-03 | | Release date: | 2006-06-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of hypothetical protein PHS023 from Pyrococcus horikoshii
TO BE PUBLISHED
|
|
2ZM8
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/D370Y Mutant Complexed with 6-Aminohexanoate-dimer | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2D0W
 
 | | Crystal structure of cytochrome cL from Hyphomicrobium denitrificans | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, cytochrome cL | | Authors: | Nojiri, M, Hira, D, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of cytochrome c(L) and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins
Biochemistry, 45, 2006
|
|
4CIS
 
 | | Structure of MutM in complex with carbocyclic 8-oxo-G containing DNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, DNA, FORMAMIDOPYRIMIDIN DNA GLYCOSYLASE, ... | | Authors: | Schneider, S, Sadeghian, K, Flaig, D, Blank, I.D, Strasser, R, Stathis, D, Winnacker, M, Carell, T, Ochsenfeld, C. | | Deposit date: | 2013-12-15 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ribose-protonated DNA base excision repair: a combined theoretical and experimental study.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
2D6C
 
 | | Crystal structure of myoglobin reconstituted with iron porphycene | | Descriptor: | IMIDAZOLE, Myoglobin, PORPHYCENE CONTAINING FE | | Authors: | Hayashi, T, Murata, D, Makino, M, Sugimoto, H, Matsuo, T, Sato, H, Shiro, Y, Hisaeda, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-11 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure and peroxidase activity of myoglobin reconstituted with iron porphycene
Inorg.Chem., 45, 2006
|
|
2D5V
 
 | | Crystal structure of HNF-6alpha DNA-binding domain in complex with the TTR promoter | | Descriptor: | 5'-D(*AP*TP*TP*AP*TP*TP*GP*AP*CP*TP*TP*AP*GP*A)-3', 5'-D(*TP*CP*TP*AP*AP*GP*TP*CP*AP*AP*TP*AP*AP*T)-3', ACETATE ION, ... | | Authors: | Iyaguchi, D, Yao, M, Watanabe, N, Nishihira, J, Tanaka, I. | | Deposit date: | 2005-11-07 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA recognition mechanism of the ONECUT homeodomain of transcription factor HNF-6
Structure, 15, 2007
|
|
