4G4L
 
 | | Crystal structure of the de novo designed peptide alpha4tbA6 | | Descriptor: | ACETYL GROUP, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2012-07-16 | | Release date: | 2012-10-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.
Protein Sci., 21, 2012
|
|
4G6P
 
 | | Minimal Hairpin Ribozyme in the Precatalytic State with A38P Variation | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.641 Å) | | Cite: | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G6S
 
 | | Minimal Hairpin Ribozyme in the Transition State with A38P Variation | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G81
 
 | | Crystal structure of a hexonate dehydrogenase ortholog (target efi-506402 from salmonella enterica, unliganded structure | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-20 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a hexonate dehydrogenase ortholog (target efi-506402 from salmonella enterica, unliganded structure
To be Published
|
|
4G8O
 
 | | Crystal Structure of a novel small molecule inactivator bound to plasminogen activator inhibitor-1 | | Descriptor: | (2S)-3-({[3-(trifluoromethyl)phenoxy]carbonyl}amino)propane-1,2-diyl bis(3,4,5-trihydroxybenzoate), 1,2-ETHANEDIOL, Plasminogen activator inhibitor 1, ... | | Authors: | Stuckey, J.A, Lawrence, D.A, Li, S.-H. | | Deposit date: | 2012-07-23 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanistic characterization and crystal structure of a small molecule inactivator bound to plasminogen activator inhibitor-1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FT7
 
 | | Crystal Structure of the CHK1 | | Descriptor: | 1-{5-bromo-2-[(3R)-3-hydroxypiperidin-1-yl]phenyl}-3-(5-cyanopyrazin-2-yl)urea, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FTJ
 
 | | Crystal Structure of the CHK1 | | Descriptor: | ISOPROPYL ALCOHOL, SULFATE ION, Serine/threonine-protein kinase Chk1, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FTS
 
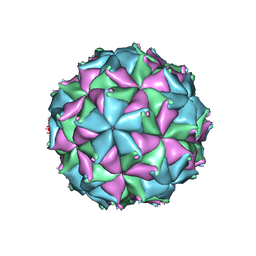 | | Crystal structure of the N363T mutant of the Flock House virus capsid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4G36
 
 | |
4GBP
 
 | | Crystal Structure of BBBB+UDP+Gal at pH 10 with MPD as the cryoprotectant | | Descriptor: | Histo-blood group ABO system transferase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Johal, A.R, Alfaro, J.A, Blackler, R.J, Schuman, B, Borisova, S.N, Evans, S.V. | | Deposit date: | 2012-07-27 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | pH-induced conformational changes in human ABO(H) blood group glycosyltransferases confirm the importance of electrostatic interactions in the formation of the semi-closed state.
Glycobiology, 24, 2014
|
|
4GAJ
 
 | | Structure of the broadly neutralizing antibody AP33 in complex with its HCV epitope (E2 residues 411-424) | | Descriptor: | Genome polyprotein, NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, NEUTRALIZING ANTIBODY AP33 LIGHT CHAIN | | Authors: | Potter, J.A, Owsianka, A, Taylor, G.L, Patel, A.H. | | Deposit date: | 2012-07-25 | | Release date: | 2012-10-10 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
1NIW
 
 | | Crystal structure of endothelial nitric oxide synthase peptide bound to calmodulin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nitric-oxide synthase, ... | | Authors: | Aoyagi, M, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-12-26 | | Release date: | 2003-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for endothelial nitric oxide synthase binding to calmodulin
Embo J., 22, 2003
|
|
4GKB
 
 | | Crystal structure of a short chain dehydrogenase homolog (target efi-505321) from burkholderia multivorans, unliganded structure | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier protein] reductase, CALCIUM ION, ... | | Authors: | Vetting, M.W, Hobbs, M.E, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a short chain dehydrogenase homolog (target efi-505321) from burkholderia multivorans, unliganded structure
To be Published
|
|
4GGH
 
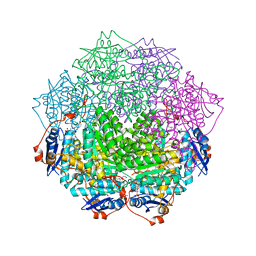 | | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, hepes, and ethylene glycol bound (ordered loops, space group c2221) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, hepes, and ethylene glycol bound (ordered loops, space group c2221)
To be Published
|
|
4FSQ
 
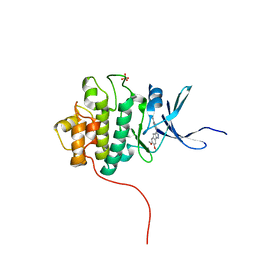 | | Crystal Structure of the CHK1 | | Descriptor: | 4'-(6,7-dimethoxyindeno[1,2-c]pyrazol-3-yl)biphenyl-4-ol, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FT0
 
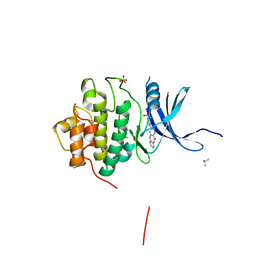 | | Crystal Structure of the CHK1 | | Descriptor: | 2-methoxy-4-(11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-3-yl)benzoic acid, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FTT
 
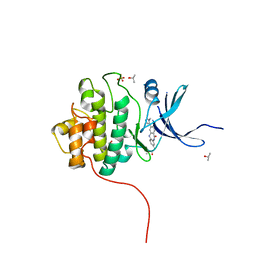 | | Crystal Structure of the CHK1 | | Descriptor: | ISOPROPYL ALCOHOL, SULFATE ION, Serine/threonine-protein kinase Chk1, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
1NBQ
 
 | | Crystal Structure of Human Junctional Adhesion Molecule Type 1 | | Descriptor: | Junctional adhesion molecule 1 | | Authors: | Prota, A.E, Campbell, J.A, Schelling, P, Forrest, J.C, Watson, M.J, Peters, T.R, Aurrand-Lions, M, Imhof, B.A, Dermody, T.S, Stehle, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human junctional adhesion molecule 1: Implications for reovirus binding
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NHC
 
 | | Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | van Pouderoyen, G, Snijder, H.J, Benen, J.A, Dijkstra, B.W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger.
Febs Lett., 554, 2003
|
|
1N7S
 
 | | High Resolution Structure of a Truncated Neuronal SNARE Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, SNAP-25A, ... | | Authors: | Ernst, J.A, Brunger, A.T. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High Resolution Structure, Stability, and Synaptotagmin Binding of a Truncated Neuronal SNARE Complex
J.Biol.Chem., 278, 2003
|
|
2W5U
 
 | | Flavodoxin from Helicobacter pylori in complex with the C3 inhibitor | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, [2-(5-amino-4-cyano-1H-pyrazol-1-yl)-5-(trifluoromethyl)phenyl](hydroxy)oxoammonium | | Authors: | Cremades, N, Perez-Dorado, I, Hermoso, J.A, Martinez-Julvez, M, Sancho, J. | | Deposit date: | 2008-12-12 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of Specific Flavodoxin Inhibitors as Potential Therapeutic Agents Against Helicobacter Pylori Infection.
Acs Chem.Biol., 4, 2009
|
|
1ZZR
 
 | | Rat nNOS D597N/M336V double mutant with L-N(omega)-Nitroarginine-(4R)-amino-L-proline amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, L-N(OMEGA)-NITROARGININE-(4R)-AMINO-L-PROLINE AMIDE, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
1ZZT
 
 | | Bovine eNOS N368D/V106M double mutant with L-N(omega)-Nitroarginine-(4R)-Amino-L-Proline Amide Bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, L-N(OMEGA)-NITROARGININE-(4R)-AMINO-L-PROLINE AMIDE, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
1ZZU
 
 | | Rat nNOS D597N/M336V double mutant with L-N(omega)-Nitroarginine-2,4-L-Diaminobutyric Amide Bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
1XT5
 
 | | Crystal Structure of VCBP3, domain 1, from Branchiostoma floridae | | Descriptor: | SULFATE ION, variable region-containing chitin-binding protein 3 | | Authors: | Hernandez Prada, J.A, Haire, R.N, Cannon, J.P, Allaire, M, Jakoncic, J, Stojanoff, V, Litman, G.W, Ostrov, D.A. | | Deposit date: | 2004-10-21 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ancient evolutionary origin of diversified variable regions demonstrated by crystal structures of an immune-type receptor in amphioxus.
Nat.Immunol., 7, 2006
|
|
