7VJ0
 
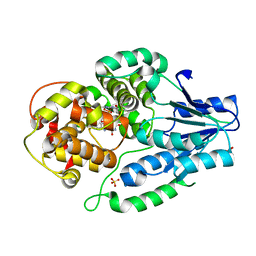 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (1 us time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJA
 
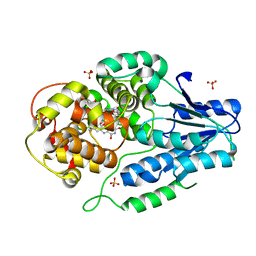 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (10 ns time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VIX
 
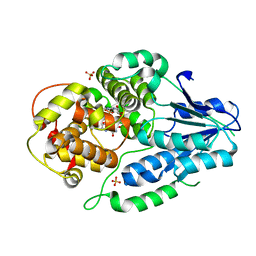 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (10 ns time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJJ
 
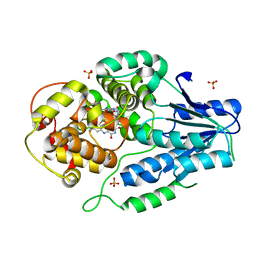 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (30 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
4UFQ
 
 | | Structure of a novel Hyaluronidase (Hyal_Sk) from Streptomyces koganeiensis. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gavira, J.A, Messina, L, Pernagallo, S, Unciti-Broceta, J.D, Conejero-Muriel, M, Diaz-Mochon, J.J, Vaccaro, S, Caruso, S, Musumeci, L, Bisicchia, S, Di Pasquale, R. | | Deposit date: | 2015-03-18 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification and Characterization of a Bacterial Hyaluronidase and its Production in Recombinant Form.
FEBS Lett., 590, 2016
|
|
5L3J
 
 | | ESCHERICHIA COLI DNA GYRASE B IN COMPLEX WITH BENZOTHIAZOLE-BASED INHIBITOR | | Descriptor: | 2-[[2-[[4,5-bis(bromanyl)-1~{H}-pyrrol-2-yl]carbonylamino]-1,3-benzothiazol-6-yl]amino]-2-oxidanylidene-ethanoic acid, DNA gyrase subunit B, IODIDE ION | | Authors: | Gjorgjieva, M, Tomasic, T, Barancokova, M, Katsamakas, S, Ilas, J, Montalvao, S, Tammella, P, Peterlin Masic, L, Kikelj, D. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Benzothiazole Scaffold-Based DNA Gyrase B Inhibitors.
J.Med.Chem., 59, 2016
|
|
7TBL
 
 | | Composite structure of the human nuclear pore complex (NPC) cytoplasmic face generated with a 12A cryo-ET map of the purified HeLa cell NPC | | Descriptor: | DDX19, ELYS, GLE1, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
6YXX
 
 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6GA7
 
 | | BACTERIORHODOPSIN, 240FS STATE, REAL-SPACE REFINED AGAINST 10% EXTRAPOLATED MAP | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
4USE
 
 | | Human STK10 (LOK) with SB-633825 | | Descriptor: | 4-{5-(6-methoxynaphthalen-2-yl)-1-methyl-2-[2-methyl-4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl}pyridine, SERINE/THREONINE-PROTEIN KINASE 10 | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Canning, P, Raynor, J, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-07-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
6GAI
 
 | | BACTERIORHODOPSIN, 740 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
5HDC
 
 | |
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
5HDD
 
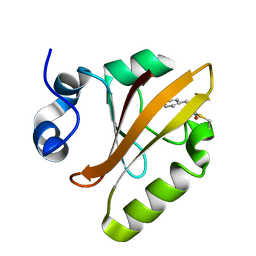 | |
4WBQ
 
 | | Crystal structure of the exonuclease domain of QIP (QDE-2 interacting protein) solved by native-SAD phasing. | | Descriptor: | CALCIUM ION, QDE-2-interacting protein | | Authors: | Boland, A, Weinert, T, Weichenrieder, O, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
5L0A
 
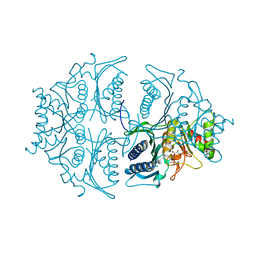 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
8PKE
 
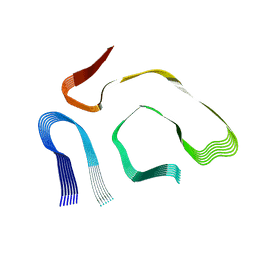 | |
8PKG
 
 | |
5L6O
 
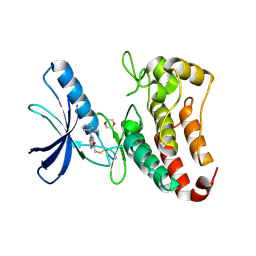 | | EphB3 kinase domain covalently bound to an irreversible inhibitor (compound 3) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 1-(4-phenylazanylquinazolin-7-yl)ethanone, Ephrin type-B receptor 3 | | Authors: | Schimpl, M, Overman, R, Kung, A, Chen, Y.-C, Ni, F, Zhu, J, Turner, M, Molina, H, Zhang, C. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Development of Specific, Irreversible Inhibitors for a Receptor Tyrosine Kinase EphB3.
J.Am.Chem.Soc., 138, 2016
|
|
7BHS
 
 | | Crystal structure of MAT2a with quinazoline fragment 2 bound in the allosteric site | | Descriptor: | 6-chloranyl-2-methoxy-4-phenyl-quinazoline, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
1JIF
 
 | | Crystal structure of bleomycin-binding protein from bleomycin-producing Streptomyces verticillus complexed with copper(II)-bleomycin | | Descriptor: | BLEOMYCIN A2, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Sugiyama, M, Kumagai, T, Hayashida, M, Maruyama, M, Matoba, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2002-02-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus.
J.Biol.Chem., 277, 2002
|
|
7BHU
 
 | | Crystal structure of MAT2a with elaborated fragment 26 bound in the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 7-chloranyl-4-(dimethylamino)-1-(2-hydroxyethyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
5HDS
 
 | |
6QGN
 
 | | Crystal structure of APT1 bound to 2-Bromopalmitate | | Descriptor: | 2-Bromopalmitic acid, Acyl-protein thioesterase 1 | | Authors: | Marcaida, M.J, Audagnotto, M, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
5FT6
 
 | | Crystal structure of the cysteine desulfurase CsdA (S-sulfonic acid) from Escherichia coli at 2.050 Angstroem resolution | | Descriptor: | CYSTEINE DESULFURASE CSDA, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
