4TZA
 
 | |
6X45
 
 | |
5XMQ
 
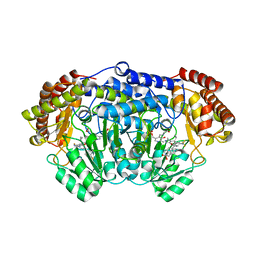 | | Plasmodium vivax SHMT(C346A) bound with PLP-glycine and MF011 | | Descriptor: | 4-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]-~{N}-methyl-~{N}-(phenylmethyl)benzenesulfonamide, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
2FME
 
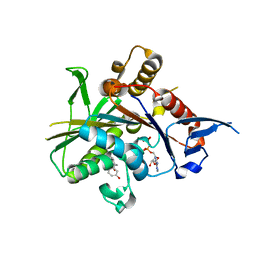 | | Crystal structure of the mitotic kinesin eg5 (ksp) in complex with mg-adp and (r)-4-(3-hydroxyphenyl)-n,n,7,8-tetramethyl-3,4-dihydroisoquinoline-2(1h)-carboxamide | | Descriptor: | (4R)-4-(3-HYDROXYPHENYL)-N,N,7,8-TETRAMETHYL-3,4-DIHYDROISOQUINOLINE-2(1H)-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Sheriff, S. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of human mitotic kinesin Eg5: Characterization of the 4-phenyl-tetrahydroisoquinoline lead series
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5XMP
 
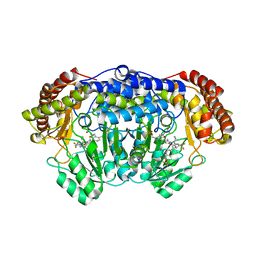 | | Plasmodium vivax SHMT(C346A) bound with PLP-glycine and MF057 | | Descriptor: | 4-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]-~{N}-methyl-~{N}-[2,2,2-tris(fluoranyl)ethyl]benzenesulfonamide, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5XMS
 
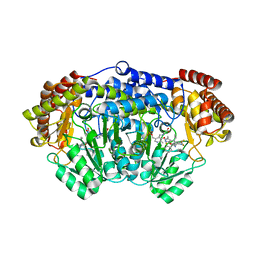 | | Plasmodium vivax SHMT bound with PLP-glycine and GS498 | | Descriptor: | (4~{S})-6-azanyl-4-[3-(2-fluorophenyl)-5-(trifluoromethyl)phenyl]-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazole-5- carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
7PUO
 
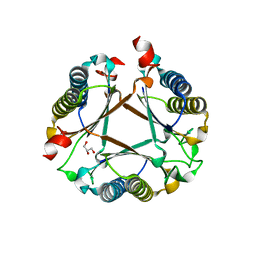 | | Structure of a fused 4-OT variant engineered for asymmetric Michael addition reactions | | Descriptor: | 2-hydroxymuconate tautomerase,Chains: A,B,C,D,E,F,2-hydroxymuconate tautomerase, CHLORIDE ION, GLYCEROL | | Authors: | Rozeboom, H.J, Thunnissen, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gene Fusion and Directed Evolution to Break Structural Symmetry and Boost Catalysis by an Oligomeric C-C Bond-Forming Enzyme.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6PRL
 
 | |
6PYQ
 
 | |
6PZ6
 
 | |
6QSQ
 
 | |
8BYP
 
 | |
5DWU
 
 | | Beta common receptor in complex with a Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, Fab - Heavy Chain, ... | | Authors: | Dhagat, U, Parker, M.W. | | Deposit date: | 2015-09-23 | | Release date: | 2015-12-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | CSL311, a novel, potent, therapeutic monoclonal antibody for the treatment of diseases mediated by the common beta chain of the IL-3, GM-CSF and IL-5 receptors.
Mabs, 8, 2016
|
|
5EML
 
 | |
5EMK
 
 | |
5EMJ
 
 | | Crystal structure of PRMT5:MEP50 with Compound 8 and sinefungin | | Descriptor: | (2~{S})-1-(3,4-dihydro-1~{H}-isoquinolin-2-yl)-3-[[4-(3-methylbenzimidazol-5-yl)pyridin-2-yl]amino]propan-2-ol, GLYCEROL, Methylosome protein 50, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2015-11-06 | | Release date: | 2016-02-24 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structure and Property Guided Design in the Identification of PRMT5 Tool Compound EPZ015666.
ACS Med Chem Lett, 7, 2016
|
|
8SGT
 
 | |
8SGJ
 
 | |
5FUN
 
 | | Crystal structure of human JARID1B in complex with GSK467 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(1-benzyl-1H-pyrazol-4-yl)oxy]pyrido[3,4-d]pyrimidin-4(3H)-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Brennan, P, Oppermann, U. | | Deposit date: | 2016-01-28 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FUP
 
 | | Crystal structure of human JARID1B in complex with 2-oxoglutarate. | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Talon, R, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FPU
 
 | | Crystal structure of human JARID1B in complex with GSKJ1 | | Descriptor: | 1,2-ETHANEDIOL, 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Nowak, R, Johansson, C, Gileadi, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2015-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FWJ
 
 | | Crystal structure of human JARID1C in complex with KDM5-C49 | | Descriptor: | 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, HISTONE DEMETHYLASE JARID1C, MAGNESIUM ION, ... | | Authors: | Srikannathasan, V, Szykowska, A, Strain-Damerell, C, Kopec, J, Nowak, R, Gileadi, C, Johansson, C, Kupinska, K, Burgess-Brown, N.A, Shrestha, L, Dong, W, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Huber, K, Oppermann, U. | | Deposit date: | 2016-02-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FV3
 
 | | Crystal structure of human JARID1B construct c2 in complex with N- Oxalylglycine. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Talon, R, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-02-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
7JSE
 
 | | Adeno-Associated Virus Origin Binding Domain in complex with ssDNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSH
 
 | | Adeno-Associated Virus 2 Rep68 HD Heptamer-ssAAVS1 with ATPgS | | Descriptor: | DNA (5'-D(P*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*CP*TP*CP*GP*C)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
