6H2Q
 
 | | Crystal Structure of Arg184Gln mutant of Human Prolidase with Mn ions and LeuPro ligand | | Descriptor: | GLYCEROL, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilk, P, Piwowarczyk, R, Weiss, M.S. | | Deposit date: | 2018-07-14 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
1FCQ
 
 | | CRYSTAL STRUCTURE (MONOCLINIC) OF BEE VENOM HYALURONIDASE | | Descriptor: | HYALURONOGLUCOSAMINIDASE | | Authors: | Markovic-Housley, Z, Miglierini, G, Soldatova, L, Mueller, U, Schirmer, T. | | Deposit date: | 2000-07-19 | | Release date: | 2001-10-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of hyaluronidase, a major allergen of bee venom.
Structure Fold.Des., 8, 2000
|
|
1FCV
 
 | | CRYSTAL STRUCTURE OF BEE VENOM HYALURONIDASE IN COMPLEX WITH HYALURONIC ACID TETRAMER | | Descriptor: | HYALURONOGLUCOSAMINIDASE, alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid | | Authors: | Markovic-Housley, Z, Miglierini, G, Soldatova, L, Rizkallah, P.J, Mueller, U, Schirmer, T. | | Deposit date: | 2000-07-19 | | Release date: | 2001-10-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of hyaluronidase, a major allergen of bee venom.
Structure Fold.Des., 8, 2000
|
|
1FCU
 
 | | CRYSTAL STRUCTURE (TRIGONAL) OF BEE VENOM HYALURONIDASE | | Descriptor: | HYALURONOGLUCOSAMINIDASE | | Authors: | Markovic-Housley, Z, Miglierini, G, Soldatova, L, Rizkallah, P.J, Mueller, U, Schirmer, T. | | Deposit date: | 2000-07-19 | | Release date: | 2001-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of hyaluronidase, a major allergen of bee venom.
Structure Fold.Des., 8, 2000
|
|
7P9T
 
 | | Crystal structure of CD73 in complex with dCMP in the open form | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PB5
 
 | | Crystal structure of CD73 in complex with UMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PBA
 
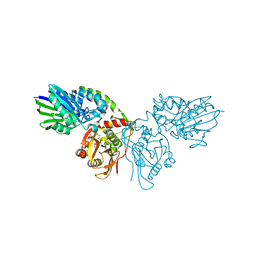 | | Crystal structure of CD73 in complex with IMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-08-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PD9
 
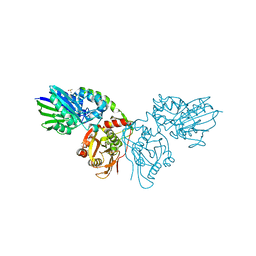 | |
7PA4
 
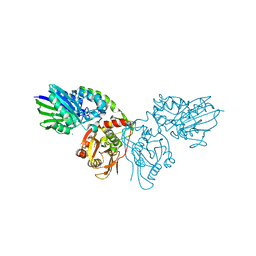 | | Crystal structure of CD73 in complex with CMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PBB
 
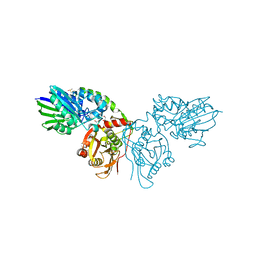 | |
7P9N
 
 | | Crystal structure of CD73 in complex with AMP in the open form | | Descriptor: | 5'-nucleotidase, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7P9R
 
 | | Crystal structure of CD73 in complex with GMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PCP
 
 | |
7PBY
 
 | |
3FGW
 
 | | One chain form of the 66.3 kDa protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, IODIDE ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
3FGR
 
 | | Two chain form of the 66.3 kDa protein at 1.8 Angstroem | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
3FGT
 
 | | Two chain form of the 66.3 kDa protein from mouse lacking the linker peptide | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
5MB7
 
 | |
5MB3
 
 | |
5MB6
 
 | |
5MB5
 
 | |
4YHU
 
 | | Yeast Prp3 C-terminal fragment 296-469 | | Descriptor: | U4/U6 small nuclear ribonucleoprotein PRP3, YTTRIUM (III) ION | | Authors: | Liu, S, Wahl, M.C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing.
Elife, 4, 2015
|
|
5QBE
 
 | | Crystal structure of Endothiapepsin-FRG175 complex | | Descriptor: | 1-[(1R)-1-cyclohexyl-2-(methylamino)-2-oxoethyl]-L-proline, ACETATE ION, Endothiapepsin, ... | | Authors: | Huschmann, F. | | Deposit date: | 2017-08-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystal structure of Endothiapepsin
To be published
|
|
5QBO
 
 | | Crystal structure of Endothiapepsin-NAT17-347283 complex | | Descriptor: | (1S,2S,3S,4R,5R)-2-[(furan-2-ylmethyl)amino]-4-(piperidin-1-yl)-6,8-dioxabicyclo[3.2.1]octan-3-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huschmann, F. | | Deposit date: | 2017-08-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | Crystal structure of Endothiapepsin
To be published
|
|
5QB6
 
 | |
