7TIX
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB56 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N~2~-{[(naphthalen-2-yl)methoxy]carbonyl}-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TJ0
 
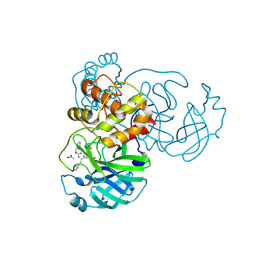 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor SL-4-241 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ACETATE ION | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7V94
 
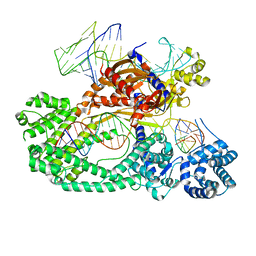 | | Cryo-EM structure of the Cas12c2-sgRNA-target DNA ternary complex | | Descriptor: | Cas12c2, sgRNA, target DNA (non target strand), ... | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
5HRT
 
 | | Crystal structure of mouse autotaxin in complex with a DNA aptamer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | Deposit date: | 2016-01-24 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis for specific inhibition of Autotaxin by a DNA aptamer
Nat.Struct.Mol.Biol., 23, 2016
|
|
7V93
 
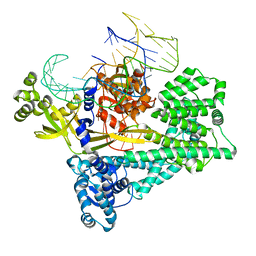 | | Cryo-EM structure of the Cas12c2-sgRNA binary complex | | Descriptor: | cas12c2, sgRNA | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
7VTI
 
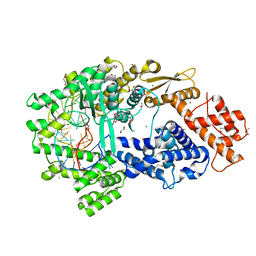 | | Crystal structure of the Cas13bt3-crRNA binary complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Nakagawa, R, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-29 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
4XZG
 
 | | Crystal structure of HIRAN domain of human HLTF | | Descriptor: | Helicase-like transcription factor | | Authors: | Ikegaya, Y, Hara, K, Hashimoto, H. | | Deposit date: | 2015-02-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Novel DNA-binding Domain of Helicase-like Transcription Factor (HLTF) and Its Functional Implication in DNA Damage Tolerance
J.Biol.Chem., 290, 2015
|
|
8GNN
 
 | | Crystal structure of the human RAD9-RAD1-HUS1-RAD17 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Cell cycle checkpoint protein RAD17, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | The 9-1-1 DNA clamp subunit RAD1 forms specific interactions with clamp loader RAD17, revealing functional implications for binding-protein RHINO.
J.Biol.Chem., 299, 2023
|
|
4YBG
 
 | | Crystal structure of the MAEL domain of Drosophila melanogaster Maelstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Protein maelstrom, ... | | Authors: | Matsumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystal Structure and Activity of the Endoribonuclease Domain of the piRNA Pathway Factor Maelstrom
Cell Rep, 11, 2015
|
|
7VTN
 
 | | Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex | | Descriptor: | Cas13bt3, crRNA, target RNA | | Authors: | Nakagawa, R, Soumya, K, Han, A, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Tomohiro, N, Yamashita, K, Feng, Z, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-30 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
3CW4
 
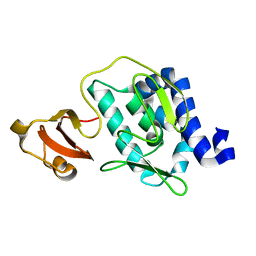 | | Large c-terminal domain of influenza a virus RNA-dependent polymerase PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Kuzuhara, T, Kise, D, Yoshida, H, Horita, T, Murasaki, Y, Utsunomiya, H, Fujiki, H, Tsuge, H. | | Deposit date: | 2008-04-21 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the influenza A virus RNA polymerase PB2 RNA-binding domain containing the pathogenicity-determinant lysine 627 residue
J.Biol.Chem., 284, 2009
|
|
6S8J
 
 | | Structure of ZEBOV GP in complex with 5T0180 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope Glycoprotein 1, Envelope glycoprotein, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-10 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
6S8I
 
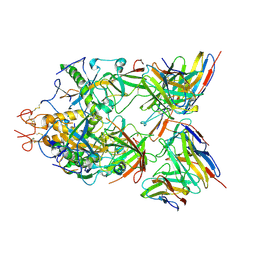 | | Structure of ZEBOV GP in complex with 3T0265 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Enveloped Glycoprotein 1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-10 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
6S9J
 
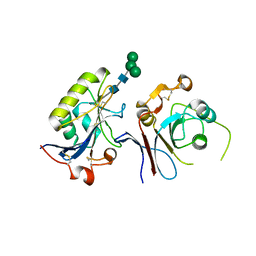 | | Crystal structure of TfR1 mimicry in complex with GP1 from MACV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pre-glycoprotein polyprotein GP complex, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-15 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Rational design of universal immunotherapy for TfR1-tropic arenaviruses.
Nat Commun, 11, 2020
|
|
6S8D
 
 | | Structure of ZEBOV GP in complex with 1T0227 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Heavy chain, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-09 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
6UWC
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-08613 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl {(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzothiazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fluorine Modifications Contribute to Potent Antiviral Activity against Highly Drug-Resistant HIV-1 and Favorable Blood-Brain Barrier Penetration Property of Novel Central Nervous System-Targeting HIV-1 Protease Inhibitors In Vitro.
Antimicrob.Agents Chemother., 66, 2022
|
|
6UWB
 
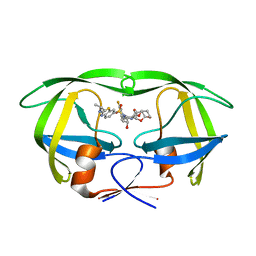 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-08513 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl {(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzothiazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fluorine Modifications Contribute to Potent Antiviral Activity against Highly Drug-Resistant HIV-1 and Favorable Blood-Brain Barrier Penetration Property of Novel Central Nervous System-Targeting HIV-1 Protease Inhibitors In Vitro.
Antimicrob.Agents Chemother., 66, 2022
|
|
5ZLG
 
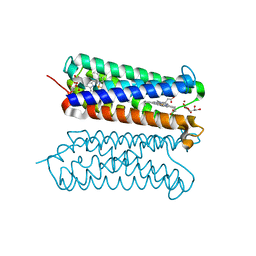 | | Human duodenal cytochrome b (Dcytb) in zinc ion and ascorbate bound form | | Descriptor: | ASCORBIC ACID, Cytochrome b reductase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ganasen, M, Togashi, H, Mauk, G.A, Shiro, Y, Sawai, H, Sugimoto, H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate.
Commun Biol, 1, 2018
|
|
8YK2
 
 | | Blood group B alpha-1,3-galactosidase AgaBb from Bifidobacterium bifidum, construct T7-tag_24-700 | | Descriptor: | Alpha-galactosidase, GLYCEROL, SODIUM ION, ... | | Authors: | Kashima, T, Akama, M, Ashida, H, Fushinobu, S. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Bifidobacterium bifidum Glycoside Hydrolase Family 110 alpha-Galactosidase Specific for Blood Group B Antigen
J.Appl.Glyosci., 2024
|
|
8YK1
 
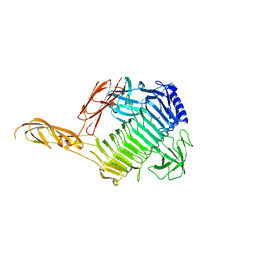 | |
8YK3
 
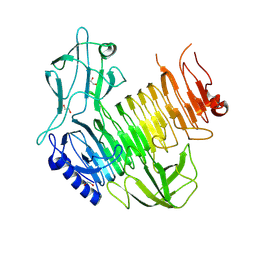 | | Blood group B alpha-1,3-galactosidase AgaBb from Bifidobacterium bifidum, construct T7-tag_24-673 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-galactosidase, ... | | Authors: | Kashima, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Bifidobacterium bifidum Glycoside Hydrolase Family 110 alpha-Galactosidase Specific for Blood Group B Antigen
J.Appl.Glyosci., 2024
|
|
4EN6
 
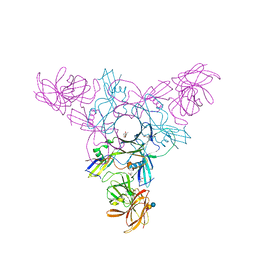 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
5JYJ
 
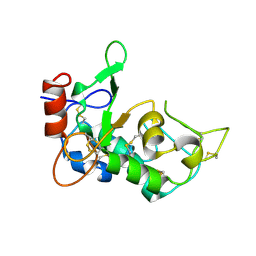 | | Crystal structure of mouse JUNO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sperm-egg fusion protein Juno | | Authors: | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the egg IZUMO1 receptor JUNO
To Be Published
|
|
4EN7
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN8
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
