2H76
 
 | |
2H6Y
 
 | |
2H71
 
 | |
4BA7
 
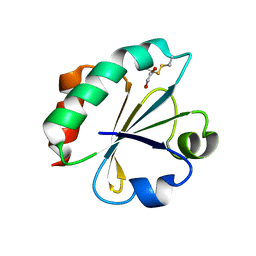 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Bacteria Common Ancestor (LBCA) from the Precambrian Period | | Descriptor: | 1,2-ETHANEDIOL, LBCA THIOREDOXIN | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-09-12 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
3ZIV
 
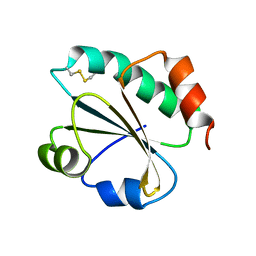 | |
3ZHC
 
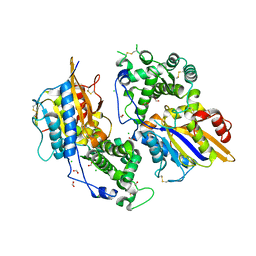 | | Structure of the phytase from Citrobacter braakii at 2.3 angstrom resolution. | | Descriptor: | CHLORIDE ION, FORMIC ACID, PHYTASE | | Authors: | Wilson, K.S, Ariza, A, Sanchez-Romero, I, Skjot, M, Vind, J, DeMaria, L, Skov, L.K, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Protein Kinetic Stabilization by Engineered Disulfide Crosslinks
Plos One, 8, 2013
|
|
4B88
 
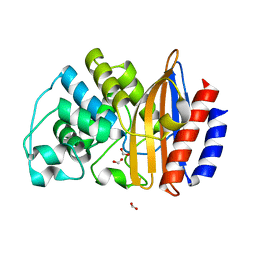 | |
3ZDJ
 
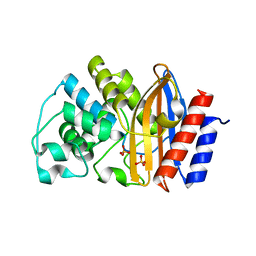 | |
2FCH
 
 | |
2H72
 
 | |
2H6Z
 
 | | Crystal Structure of Thioredoxin Mutant E44D in Hexagonal (p61) Space Group | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thioredoxin | | Authors: | Gavira, J.A, Godoy-Ruiz, R, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A stability pattern of protein hydrophobic mutations that reflects evolutionary structural optimization.
Biophys.J., 89, 2005
|
|
2H70
 
 | | Crystal Structure of Thioredoxin Mutant D9E in Hexagonal (p61) Space Group | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thioredoxin | | Authors: | Gavira, J.A, Godoy-Ruiz, R, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A stability pattern of protein hydrophobic mutations that reflects evolutionary structural optimization.
Biophys.J., 89, 2005
|
|
2FD3
 
 | |
6QIZ
 
 | | CI-2, conformation 2 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
6QIY
 
 | | CI-2, conformation 1 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
2YJ7
 
 | | Crystal structure of a hyperstable protein from the Precambrian period | | Descriptor: | LPBCA THIOREDOXIN, SODIUM ION | | Authors: | Gavira, J.A, Ingles, A, Ibarra, B, Garcia-Ruiz, J.M. | | Deposit date: | 2011-05-19 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
5FQM
 
 | | Last common ancestor of Gram Negative Bacteria (GNCA) Class A beta- lactamase | | Descriptor: | GLYCEROL, GNCA BETA LACTAMASE, SULFATE ION | | Authors: | Martinez Rodriguez, S, Gavira, J.A, Risso, V.A, Sanchez Ruiz, J.M. | | Deposit date: | 2015-12-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
4JEQ
 
 | | Different Contribution of Conserved Amino Acids to the Global Properties of Homologous Enzymes | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Hernandez-Santoyo, A, Aguirre-Fuentes, Y, Torres-Larios, A, Gomez-Puyou, A, De Gomez-Puyou, M.T. | | Deposit date: | 2013-02-27 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Different contribution of conserved amino acids to the global properties of triosephosphate isomerases.
Proteins, 82, 2014
|
|
4HHP
 
 | | Crystal structure of triosephosphate isomerase from trypanosoma cruzi, mutant e105d | | Descriptor: | GLYCEROL, SULFATE ION, Triosephosphate isomerase, ... | | Authors: | Hernandez-Santoyo, A, Aguirre-Fuentes, Y, Torres-Larios, A, Gomez-Puyou, A, De Gomez-Puyou, M.T. | | Deposit date: | 2012-10-10 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Different contribution of conserved amino acids to the global properties of triosephosphate isomerases.
Proteins, 82, 2014
|
|
1R4Y
 
 | | SOLUTION STRUCTURE OF THE DELETION MUTANT DELTA(7-22) OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | Ribonuclease alpha-sarcin | | Authors: | Garcia-Mayoral, M.F, Garcia-Ortega, L, Lillo, M.P, Santoro, J, Martinez Del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 2003-10-09 | | Release date: | 2004-04-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the noncytotoxic {alpha}-sarcin mutant {Delta}(7-22): The importance of the native conformation of peripheral loops for activity.
Protein Sci., 13, 2004
|
|
4CBR
 
 | | X-ray structure of the more stable human AGXT triple mutant (AGXT_HEM) | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, SERINE--PYRUVATE AMINOTRANSFERASE | | Authors: | Yunta, C, Albert, A. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Consensus-Based Approach for Gene/Enzyme Replacement Therapies and Crystallization Strategies: The Case of Human Alanine:Glyoxylate Aminotransferase.
Biochem.J., 462, 2014
|
|
4CBS
 
 | |
