7ASV
 
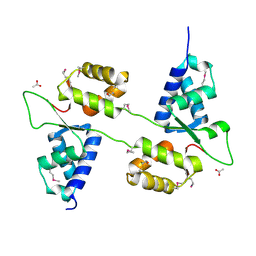 | |
7ASU
 
 | |
1GVL
 
 | | Human prokallikrein 6 (hK6)/ prozyme/ proprotease M/ proneurosin | | Descriptor: | KALLIKREIN 6 | | Authors: | Gomis-Ruth, F.X, Bayes, A, Sotiropoulou, G, Pampalakis, G, Tsetsenis, T, Villegas, V, Aviles, F.X, Coll, M. | | Deposit date: | 2002-02-14 | | Release date: | 2002-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Human Prokallikrein 6 Reveals a Novel Activation Mechanism for the Kallikrein Family.
J.Biol.Chem., 277, 2002
|
|
8A43
 
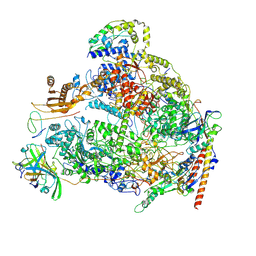 | | Human RNA polymerase I | | Descriptor: | DNA-directed RNA polymerase I subunit RPA1, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA2, ... | | Authors: | Daiss, J.L, Pilsl, M, Straub, K, Bleckmann, A, Hoecherl, M, Heiss, F.B, Abascal-Palacios, G, Ramsay, E, Tluckova, K, Mars, J.C, Fuertges, T, Bruckmann, A, Rudack, T, Bernecky, C, Lamour, V, Panov, K, Vannini, A, Moss, T, Engel, C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-31 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | The human RNA polymerase I structure reveals an HMG-like docking domain specific to metazoans.
Life Sci Alliance, 5, 2022
|
|
5VOE
 
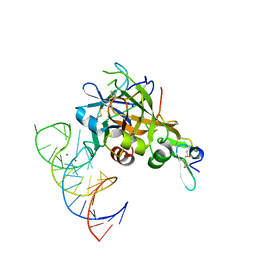 | | DesGla-XaS195A Bound to Aptamer 11F7t | | Descriptor: | Aptamer 11F7t (36-MER), CALCIUM ION, Coagulation factor X, ... | | Authors: | Gunaratne, R, Kumar, S, Frederiksen, J.W, Stayrook, S, Lohrmann, J.L, Perry, K, Chabata, C.V, Thalji, N.K, Ho, M.D, Arepally, G, Camire, R.M, Krishnaswamy, S.K, Sullenger, B.A. | | Deposit date: | 2017-05-02 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass.
Nat. Biotechnol., 36, 2018
|
|
1IF8
 
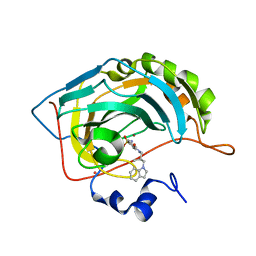 | | Carbonic Anhydrase II Complexed With (S)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | Descriptor: | (S)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IF7
 
 | | Carbonic Anhydrase II Complexed With (R)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | Descriptor: | (R)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IF9
 
 | | Carbonic Anhydrase II Complexed With N-[2-(1H-Indol-5-yl)-butyl]-4-sulfamoyl-benzamide | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-[2-(1H-INDOL-5-YL)-BUTYL]-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Y0O
 
 | |
1EMM
 
 | |
1EML
 
 | |
1EMC
 
 | |
1EMK
 
 | |
1EME
 
 | |
1EMF
 
 | |
2EMN
 
 | |
2EMD
 
 | |
2EMO
 
 | |
7PQO
 
 | | Catalytic fragment of MASP-1 in complex with P1 site mutant ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 1, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
7PQN
 
 | | Catalytic fragment of MASP-2 in complex with ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 2 A chain, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.400015 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
7ARX
 
 | | Crystal structure of the catalytic fragment of masp-1 in complex with SFMI1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Mannan-binding lectin serine protease 1, SFMI1 - Sunflower MASP1 inhibitor | | Authors: | Durvanger, Z, Harmat, V, Dobo, J, Megyeri, M. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Directed Evolution-Driven Increase of Structural Plasticity Is a Prerequisite for Binding the Complement Lectin Pathway Blocking MASP-Inhibitor Peptides.
Acs Chem.Biol., 17, 2022
|
|
2B59
 
 | |
4IGD
 
 | | Crystal structure of the zymogen catalytic region of Human MASP-1 | | Descriptor: | GLYCEROL, Mannan-binding lectin serine protease 1 | | Authors: | Harmat, V, Megyeri, M, Vegh, A, Dobo, J. | | Deposit date: | 2012-12-17 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative characterization of the activation steps of mannan-binding lectin (MBL)-associated serine proteases (MASPs) points to the central role of MASP-1 in the initiation of the complement lectin pathway
J.Biol.Chem., 288, 2013
|
|
6GW9
 
 | | Concanavalin A structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | CALCIUM ION, Concanavalin V, MAGNESIUM ION | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6GWA
 
 | | Concanavalin B structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | Concanavalin B | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
