6M2E
 
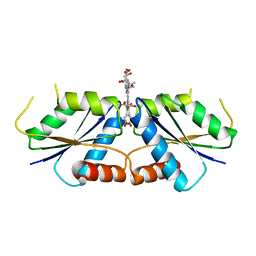 | | Sirohydrochlorin nickelochelatase CfbA in complex with sirohydrochlorin | | Descriptor: | 3,3',3'',3'''-[(7S,8S,12S,13S)-3,8,13,17-tetrakis(carboxymethyl)-8,13-dimethyl-7,8,12,13-tetrahydroporphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, Sirohydrochlorin cobaltochelatase | | Authors: | Fujishiro, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The nickel-sirohydrochlorin formation mechanism of the ancestral class II chelatase CfbA in coenzyme F430 biosynthesis.
Chem Sci, 12, 2021
|
|
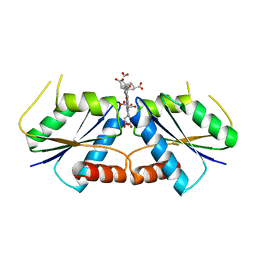
6M2F
 
 | |
6M26
 
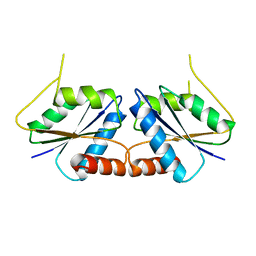 | | Sirohydrochlorin nickelochelatase CfbA in P212121 space group | | Descriptor: | Sirohydrochlorin cobaltochelatase | | Authors: | Fujishiro, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The nickel-sirohydrochlorin formation mechanism of the ancestral class II chelatase CfbA in coenzyme F430 biosynthesis.
Chem Sci, 12, 2021
|
|
6M29
 
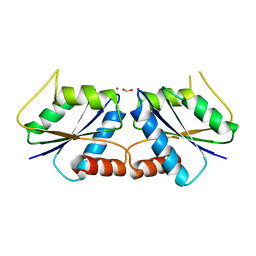 | |
6KFY
 
 | |
6KG1
 
 | | NifS from Helicobacter pylori, soaked with L-cysteine for 180 sec | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, CHLORIDE ION, Cysteine desulfurase IscS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
6KG0
 
 | | NifS from Helicobacter pylori, soaked with L-cysteine for 118 sec | | Descriptor: | CHLORIDE ION, Cysteine desulfurase IscS, ISOPROPYL ALCOHOL, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
6KFZ
 
 | | SufS from Bacillus subtilis, soaked with L-cysteine for 90 sec at 1.96 angstrom resolution | | Descriptor: | Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
5ZS9
 
 | |
5ZST
 
 | |
7CET
 
 | | Crystal structure of D-cycloserine-bound form of cysteine desulfurase NifS from Helicobacter pylori | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, CHLORIDE ION, Cysteine desulfurase IscS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEQ
 
 | |
7CER
 
 | | Crystal structure of D-cycloserine-bound form of cysteine desulfurase SufS H121A from Bacillus subtilis | | Descriptor: | Cysteine desulfurase SufS, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEP
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Yasuhiro, T, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEU
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase NifS from Helicobacter pylori | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase IscS, ISOPROPYL ALCOHOL | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEO
 
 | |
7CES
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS H121A from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7E6A
 
 | | Crystal structure of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6E
 
 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in D-cycloserine-inhibition | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6C
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6B
 
 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6F
 
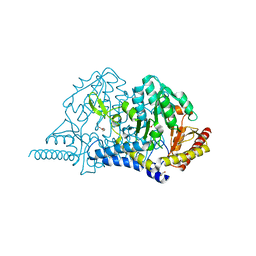 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in L-cycloserine-inhibition | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6D
 
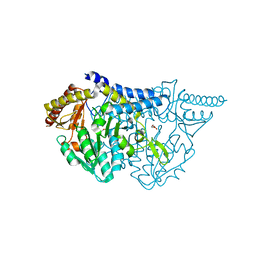 | |
