8C4U
 
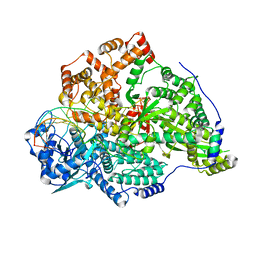 | | Hantaan virus polymerase in replication pre-initiation state | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*UP*AP*GP*GP*AP*GP*UP*AP*UP*CP*CP*AP*CP*CP*GP*CP*AP*AP*GP*A)-3'), RNA (5'-R(P*UP*UP*UP*UP*GP*CP*GP*GP*AP*GP*UP*AP*CP*UP*AP*CP*UP*A)-3'), ... | | Authors: | Durieux trouilleton, Q, Arragain, B, Malet, H. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structures of active Hantaan virus polymerase uncover the mechanisms of Hantaviridae genome replication.
Nat Commun, 14, 2023
|
|
8C4V
 
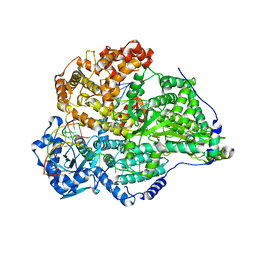 | | Hantaan virus polymerase in replication elongation state | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*CP*UP*AP*CP*UP*A)-3'), RNA (5'-R(P*AP*GP*GP*AP*GP*UP*AP*UP*CP*CP*AP*CP*CP*GP*CP*AP*AP*GP*A)-3'), ... | | Authors: | Durieux trouilleton, Q, Arragain, B, Malet, H. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structures of active Hantaan virus polymerase uncover the mechanisms of Hantaviridae genome replication.
Nat Commun, 14, 2023
|
|
8C4S
 
 | |
8QE5
 
 | |
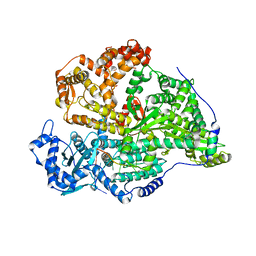
8QGT
 
 | |
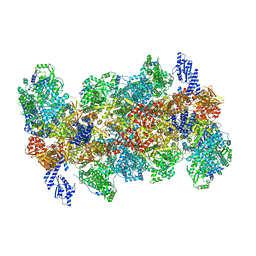
8QHD
 
 | |
8QGU
 
 | |
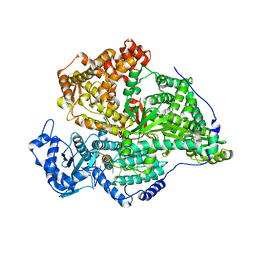
8QH3
 
 | |
2J7W
 
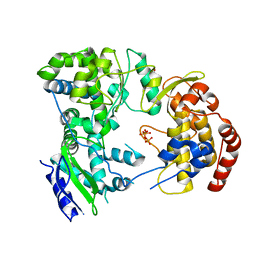 | | Dengue virus NS5 RNA dependent RNA polymerase domain complexed with 3' dGTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-TRIPHOSPHATE, POLYPROTEIN, ... | | Authors: | Yap, T.L, Xu, T, Chen, Y.L, Malet, H, Egloff, M.P, Canard, B, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Dengue Virus RNA- Dependent RNA Polymerase Catalytic Domain at 1.85 Angstrom Resolution.
J.Virol., 81, 2007
|
|
3STJ
 
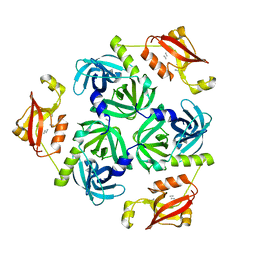 | | Crystal structure of the protease + PDZ1 domain of DegQ from Escherichia coli | | Descriptor: | Protease degQ, peptide (UNK) | | Authors: | Sawa, J, Malet, H, Krojer, T, Canellas, F, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope.
J.Biol.Chem., 286, 2011
|
|
3STI
 
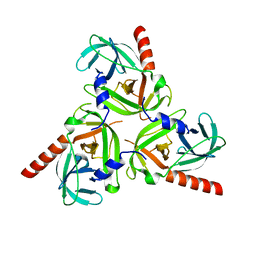 | | Crystal structure of the protease domain of DegQ from Escherichia coli | | Descriptor: | Protease degQ | | Authors: | Sawa, J, Malet, H, Krojer, T, Canellas, F, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope.
J.Biol.Chem., 286, 2011
|
|
5FKX
 
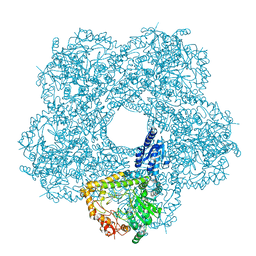 | | Structure of E.coli inducible lysine decarboxylase at active pH | | Descriptor: | LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
5FKZ
 
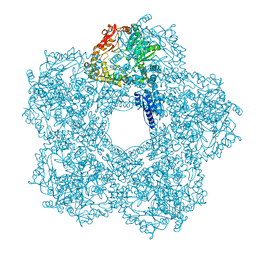 | | Structure of E.coli Constitutive lysine decarboxylase | | Descriptor: | LYSINE DECARBOXYLASE, CONSTITUTIVE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
8CI5
 
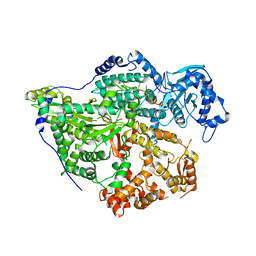 | | Structure of the SNV L protein bound to 5' RNA | | Descriptor: | RNA (5'-R(P*AP*GP*UP*AP*GP*UP*AP*GP*AP*CP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Meier, K, Thorkelsson, S.R, Durieux Trouilleton, Q, Vogel, D, Yu, D, Kosinski, J, Cusack, S, Malet, H, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-02-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of the Sin Nombre virus L protein.
Plos Pathog., 19, 2023
|
|
7ORJ
 
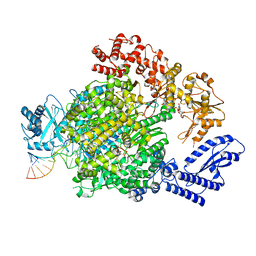 | | La Crosse virus polymerase at transcription capped RNA cleavage stage | | Descriptor: | 5' capped RNA, ADENOSINE-5'-TRIPHOSPHATE, RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*CP*AP*AP*G)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORI
 
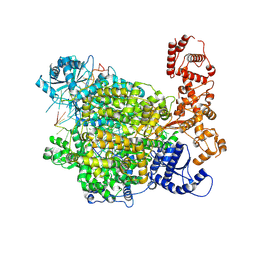 | | La Crosse virus polymerase at replication late-elongation stage | | Descriptor: | La Crosse virus polymerase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORO
 
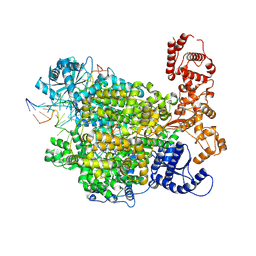 | | La Crosse virus polymerase at replication early-elongation stage | | Descriptor: | La Crosse virus polymerase, MAGNESIUM ION, RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*C)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORN
 
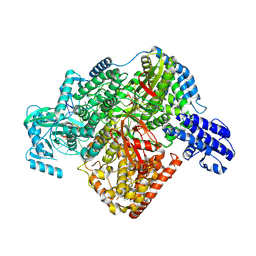 | | La Crosse virus polymerase at replication initiation stage | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, La Crosse virus polymerase, MAGNESIUM ION, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORL
 
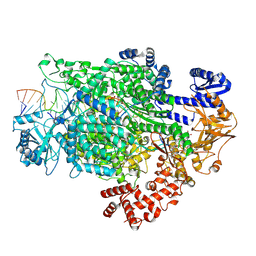 | | La Crosse virus polymerase at transcription initiation stage | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORM
 
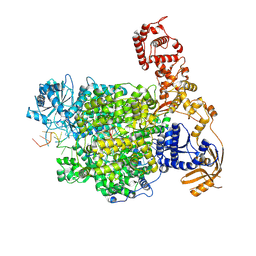 | | La Crosse virus polymerase at transcription early-elongation stage | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*GP*UP*AP*CP*AP*CP*UP*AP*CP*U)-3'), RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*C)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORK
 
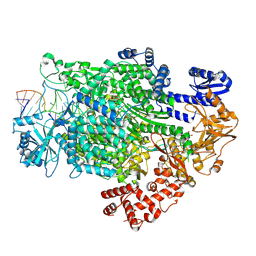 | | La Crosse virus polymerase in transcription mode with cleaved capped RNA entering the polymerase active site | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (5'-D(*(GTG))-R(P*A)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
2FAV
 
 | |
4BGP
 
 | | Crystal structure of La Crosse virus nucleoprotein | | Descriptor: | GLYCEROL, NUCLEOPROTEIN, SULFATE ION | | Authors: | Reguera, J, Malet, H, Weber, F, Cusack, S. | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Encapsidation of Genomic RNA by La Crosse Orthobunyavirus Nucleoprotein
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BHH
 
 | | Crystal structure of tetramer of La Crosse virus nucleoprotein in complex with ssRNA | | Descriptor: | NUCLEOPROTEIN, POLY-URIDINE 45-MER | | Authors: | Reguera, J, Malet, H, Weber, F, Cusack, S. | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for Encapsidation of Genomic RNA by La Crosse Orthobunyavirus Nucleoprotein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6Z6B
 
 | | Structure of full-length La Crosse virus L protein (polymerase) | | Descriptor: | RNA (5'-R(*GP*CP*UP*AP*CP*UP*AP*A)-3'), RNA (5'-R(P*AP*GP*UP*AP*GP*UP*GP*UP*GP*C)-3'), RNA (5'-R(P*UP*UP*AP*GP*UP*AP*GP*UP*AP*CP*AP*CP*UP*AP*CP*U)-3'), ... | | Authors: | Cusack, S, Gerlach, P, Reguera, J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
