2R02
 
 | |
4WLP
 
 | | Crystal structure of UCH37-NFRKB Inhibited Deubiquitylating Complex | | Descriptor: | Nuclear factor related to kappa-B-binding protein, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
4WNN
 
 | | SPT16-H2A-H2B FACT HISTONE Complex | | Descriptor: | Histone H2A.1, Histone H2B.1, PHOSPHATE ION, ... | | Authors: | Kemble, D.J, Hill, C.P, Whitby, F.G, Formosa, T, McCullough, L.L. | | Deposit date: | 2014-10-13 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | FACT Disrupts Nucleosome Structure by Binding H2A-H2B with Conserved Peptide Motifs.
Mol.Cell, 60, 2015
|
|
4WLQ
 
 | | Crystal structure of mUCH37-hRPN13 CTD complex | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
1CMX
 
 | |
3DFZ
 
 | | SirC, precorrin-2 dehydrogenase | | Descriptor: | GLYCEROL, Precorrin-2 dehydrogenase, SULFATE ION | | Authors: | Schubert, H.L, Hill, C.P, Warren, M.J. | | Deposit date: | 2008-06-12 | | Release date: | 2008-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of SirC from Bacillus megaterium: a metal-binding precorrin-2 dehydrogenase
Biochem.J., 415, 2008
|
|
1JPH
 
 | | Ile260Thr mutant of Human UroD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
1JPI
 
 | | Phe232Leu mutant of human UROD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
1JPK
 
 | | Gly156Asp mutant of Human UroD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
4WLR
 
 | | Crystal Structure of mUCH37-hRPN13 CTD-hUb complex | | Descriptor: | Polyubiquitin-B, Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
1TLB
 
 | | Yeast coproporphyrinogen oxidase | | Descriptor: | Coproporphyrinogen III oxidase, SULFATE ION | | Authors: | Phillip, J.D, Whitby, F.G, Warby, C.A, Labbe, P, Yang, C, Pflugrath, J.W, Ferrara, J.D, Robinson, H, Kushner, J.P, Hill, C.P. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the oxygen-dependent coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
1URO
 
 | | UROPORPHYRINOGEN DECARBOXYLASE | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (UROPORPHYRINOGEN DECARBOXYLASE) | | Authors: | Whitby, F.G, Phillips, J.D, Kushner, J.P, Hill, C.P. | | Deposit date: | 1998-08-21 | | Release date: | 1998-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human uroporphyrinogen decarboxylase.
EMBO J., 17, 1998
|
|
1TK1
 
 | | YEAST OXYGEN-DEPENDENT COPROPORPHYRINOGEN OXIDASE | | Descriptor: | Coproporphyrinogen III oxidase | | Authors: | Phillips, J.D, Whitby, F.G, Warby, C.A, Labbe, P, Yang, C, Pflugrath, J.W, Ferrara, J.D, Robinson, H, Kushner, J.P, Hill, C.P. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Oxygen-dependant Coproporphyrinogen Oxidase (Hem13p) of Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
1TKL
 
 | | Yeast Oxygen-Dependent Coproporphyrinogen Oxidase | | Descriptor: | Coproporphyrinogen III oxidase | | Authors: | Phillips, J.D, Whitby, F.G, Warby, C.A, Labbe, P, Yang, C, Pflugrath, J.W, Ferrara, J.D, Robinson, H, Kushner, J.P, Hill, C.P. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Oxygen-dependant Coproporphyrinogen Oxidase (Hem13p) of Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
6OMB
 
 | | Cdc48 Hexamer (Subunits A to E) with substrate bound to the central pore | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Cooney, I, Han, H, Stewart, M, Carson, R.H, Hansen, D, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Cdc48 segregase in the act of unfolding an authentic substrate.
Science, 365, 2019
|
|
6OPC
 
 | | Cdc48 Hexamer in a complex with substrate and Shp1(Ubx Domain) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Cooney, I, Han, H, Stewart, M, Carson, R.H, Hansen, D, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Cdc48 segregase in the act of unfolding an authentic substrate.
Science, 365, 2019
|
|
6OO2
 
 | | Vps4 with Cyclic Peptide Bound in the Central Pore | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Designed Cyclic Peptide, ... | | Authors: | Han, H, Fulcher, J.M, Dandey, V.P, Sundquist, W.I, Kay, M.S, Shen, P, Hill, C.P. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases.
Elife, 8, 2019
|
|
6OI4
 
 | | RPN13 (19-132)-RPN2 (940-952) pY950-Ub complex | | Descriptor: | 1,2-ETHANEDIOL, 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, ... | | Authors: | Hemmis, C.W, Hill, C.P. | | Deposit date: | 2019-04-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Phosphorylation of Tyr-950 in the proteasome scaffolding protein RPN2 modulates its interaction with the ubiquitin receptor RPN13.
J.Biol.Chem., 294, 2019
|
|
6PEK
 
 | | Structure of Spastin Hexamer (Subunit A-E) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
6PEN
 
 | | Structure of Spastin Hexamer (whole model) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, EYEYEYEYEY, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
6NDY
 
 | | Vps4 with Cyclic Peptide Bound in the Central Pore | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Designed Cyclic Peptide, ... | | Authors: | Han, H, Fulcher, J.M, Dandey, V.P, Sundquist, W.I, Kay, M.S, Shen, P, Hill, C.P. | | Deposit date: | 2018-12-14 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases.
Elife, 8, 2019
|
|
3ZXP
 
 | |
8UC6
 
 | | Calpain-7:IST1 Complex | | Descriptor: | Calpain-7, IST1 homolog | | Authors: | Paine, E, Whitby, F.G, Hill, C.P, Sunquist, W.I. | | Deposit date: | 2023-09-25 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | The Calpain-7 protease functions together with the ESCRT-III protein IST1 within the midbody to regulate the timing and completion of abscission.
Elife, 12, 2023
|
|
8V14
 
 | | Structure of NRAP-1 and its role in NMDAR signaling | | Descriptor: | CALCIUM ION, NMDA receptor auxiliary protein | | Authors: | Whitby, F.G, Goodell, D.J, Maricq, A.V, Hill, C.P. | | Deposit date: | 2023-11-19 | | Release date: | 2024-01-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and structural studies reveal NRAP-1-dependent coincident activation of NMDARs.
Cell Rep, 43, 2024
|
|
7SMU
 
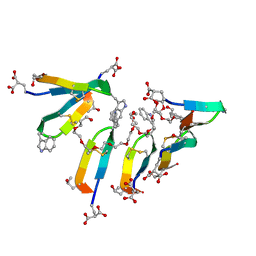 | | Crystal Structure of Consomatin-Ro1 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45-pentadecaoxaoctatetracontane-1,48-diol, Consomatin-Ro1 | | Authors: | Ramiro, I.B.L, Whitby, F.G, Hill, C.P, Safavi-Hemami, H, Concepcion, G.P, Olivera, B.M. | | Deposit date: | 2021-10-26 | | Release date: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Somatostatin venom analogs evolved by fish-hunting cone snails: From prey capture behavior to identifying drug leads.
Sci Adv, 8, 2022
|
|
