6RFG
 
 | |
2R5A
 
 | |
2R5M
 
 | |
2R57
 
 | |
2R58
 
 | |
7AMV
 
 | | Atomic structure of the poxvirus transcription pre-initiation complex in the initially melted state | | Descriptor: | ATP-dependent helicase VETFS, DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AOF
 
 | | Atomic structure of the poxvirus transcription late pre-initiation complex | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AP8
 
 | | Atomic structure of the poxvirus initially transcribing complex in conformation 2 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AOZ
 
 | | Atomic structure of the poxvirus transcription initiation complex in conformation 1 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-15 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AOH
 
 | | Atomic structure of the poxvirus late initially transcribing complex | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AP9
 
 | | Atomic structure of the poxvirus initially transcribing complex in conformation 3 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NML
 
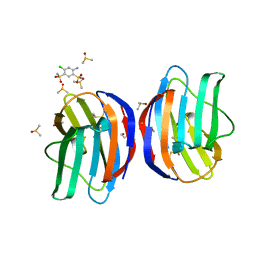 | | Galectin-1 in complex with 4-Amino-6-chloro-1,3-benzenedisulfonamide | | Descriptor: | 4-AMINO-6-CHLOROBENZENE-1,3-DISULFONAMIDE, DIMETHYL SULFOXIDE, Galectin-1 | | Authors: | Grimm, C, Bechold, J, Seibel, J. | | Deposit date: | 2021-02-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Galectin-1 in complex with 4-Amino-6-chloro-1,3-benzenedisulfonamide
To Be Published
|
|
6EK5
 
 | |
5C8B
 
 | |
4WKG
 
 | |
6RIC
 
 | | Structure of the core Vaccinia Virus DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, H.S, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
5MAN
 
 | |
5M9X
 
 | | Structure of sucrose phosphorylase from Bifidobacterium adolescentis bound to glycosylated resveratrol | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[3-[(~{E})-2-(4-hydroxyphenyl)ethenyl]-5-oxidanyl-phenoxy]oxane-3,4 ,5-triol, Sucrose phosphorylase | | Authors: | Grimm, C, Kraus, M. | | Deposit date: | 2016-11-02 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Switching enzyme specificity from phosphate to resveratrol glucosylation.
Chem. Commun. (Camb.), 53, 2017
|
|
7PLK
 
 | |
6H54
 
 | | CRYSTAL STRUCTURE OF BOVINE HSC70(AA1-554)E213A/D214A IN COMPLEX WITH INHIBITOR VER155008 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, GLYCEROL, ... | | Authors: | Plank, C, Zehe, M, Grimm, C, Sotriffer, C. | | Deposit date: | 2018-07-23 | | Release date: | 2019-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Combined In-Solution Fragment Screening and Crystallographic Binding-Mode Analysis with a Two-Domain Hsp70 Construct.
Acs Chem.Biol., 2024
|
|
8Q3R
 
 | | Cryo-EM structure of the DNA polymerase holoenzyme E9-A20-D4 of vaccinia virus | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component OPG148, Uracil-DNA glycosylase | | Authors: | Burmeister, W.P, Ballandras-Colas, A, Boettcher, B, Grimm, C. | | Deposit date: | 2023-08-04 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.
Plos Pathog., 20, 2024
|
|
1M5H
 
 | | Formylmethanofuran:tetrahydromethanopterin formyltransferase from Archaeoglobus fulgidus | | Descriptor: | Formylmethanofuran--tetrahydromethanopterin formyltransferase, POTASSIUM ION | | Authors: | Mamat, B, Roth, A, Grimm, C, Ermler, U, Tziatzios, C, Schubert, D, Thauer, R.K, Shima, S. | | Deposit date: | 2002-07-09 | | Release date: | 2002-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and enzymatic properties of three formyltransferases from archaea: environmental adaptation and evolutionary relationship.
Protein Sci., 11, 2002
|
|
2X6V
 
 | | Crystal structure of human TBX5 in the DNA-bound and DNA-free form | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5'-D(*TP*AP*AP*GP*GP*TP*GP*TP*GP*AP*GP)-3', 5'-D(*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP)-3', ... | | Authors: | Ptchelkine, D, Stirnimann, C.U, Grimm, C, Mueller, C.W. | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Tbx5-DNA Recognition: The T-Box Domain in its DNA-Bound and -Unbound Form.
J.Mol.Biol., 400, 2010
|
|
1M5S
 
 | | Formylmethanofuran:tetrahydromethanopterin fromyltransferase from Methanosarcina barkeri | | Descriptor: | Formylmethanofuran--tetrahydromethanopterin formyltransferase | | Authors: | Mamat, B, Roth, A, Grimm, C, Ermler, U, Tziatzios, C, Schubert, D, Thauer, R.K, Shima, S. | | Deposit date: | 2002-07-10 | | Release date: | 2002-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and enzymatic properties of three formyltransferases from archaea: environmental adaptation and evolutionary relationship.
Protein Sci., 11, 2002
|
|
8APL
 
 | | Vaccinia virus DNA helicase D5 residues 323-785 hexamer with bound DNA processed in C6 | | Descriptor: | Primase D5 | | Authors: | Burmeister, W.P, Hutin, S, Ling, W.L, Grimm, C, Schoehn, G. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The Vaccinia Virus DNA Helicase Structure from Combined Single-Particle Cryo-Electron Microscopy and AlphaFold2 Prediction.
Viruses, 14, 2022
|
|
