3K1Q
 
 | | Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics | | Descriptor: | Core protein VP6, Outer capsid VP5, Outer capsid VP7, ... | | Authors: | Cheng, L.P, Zhu, J, Hiu, W.H, Zhang, X.K, Honig, B, Fang, Q, Zhou, Z.H. | | Deposit date: | 2009-09-28 | | Release date: | 2010-03-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Backbone Model of an Aquareovirus Virion by Cryo-Electron Microscopy and Bioinformatics
J.Mol.Biol., 397, 2010
|
|
7LCH
 
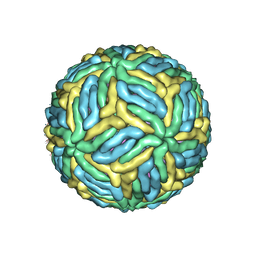 | | The mature Usutu SAAR-1776, Model B | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Khare, B, Klose, T, Fang, Q, Kuhn, R. | | Deposit date: | 2021-01-11 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structure of Usutu virus SAAR-1776 displays fusion loop asymmetry.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LCG
 
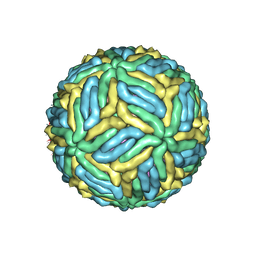 | | The mature Usutu SAAR-1776, Model A | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Khare, B, Klose, T, Fang, Q, Kuhn, R. | | Deposit date: | 2021-01-11 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structure of Usutu virus SAAR-1776 displays fusion loop asymmetry.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5ZVS
 
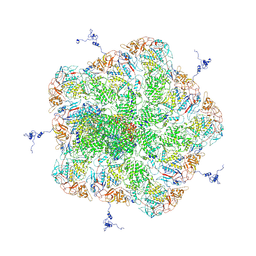 | |
5ZVT
 
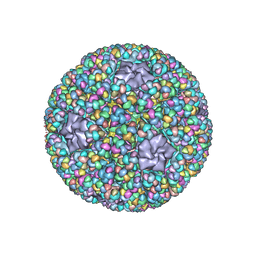 | | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly | | Descriptor: | C-terminus of outer capsid protein VP5, Core protein VP6, MYRISTIC ACID, ... | | Authors: | Liu, H, Fang, Q, Cheng, L. | | Deposit date: | 2018-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4KUI
 
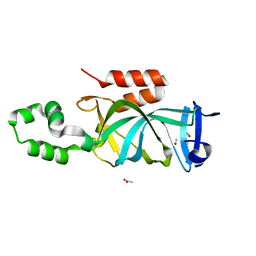 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8Y6V
 
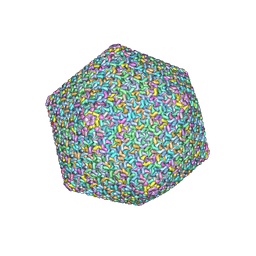 | | Near-atomic structure of icosahedrally averaged jumbo bacteriophage PhiKZ capsid | | Descriptor: | gp119, gp120, gp162, ... | | Authors: | Yang, Y, Shao, Q, Guo, M, Han, L, Zhao, X, Wang, A, Li, X, Wang, B, Pan, J, Chen, Z, Fokine, A, Sun, L, Fang, Q. | | Deposit date: | 2024-02-03 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capsid structure of bacteriophage Phi KZ provides insights into assembly and stabilization of jumbo phages.
Nat Commun, 15, 2024
|
|
4KUL
 
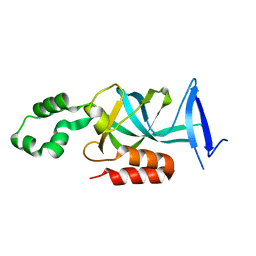 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | Descriptor: | Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8H2I
 
 | | Near-atomic structure of five-fold averaged PBCV-1 capsid | | Descriptor: | MCPv1, MCPv2, MCPv3, ... | | Authors: | Shao, Q, Agarkova, I.V, Noel, E.A, Dunigan, D.D, Liu, Y, Wang, A, Guo, M, Xie, L, Zhao, X, Rossmann, M.G, Van Etten, J.L, Klose, T, Fang, Q. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Near-atomic, non-icosahedrally averaged structure of giant virus Paramecium bursaria chlorella virus 1.
Nat Commun, 13, 2022
|
|
8IZD
 
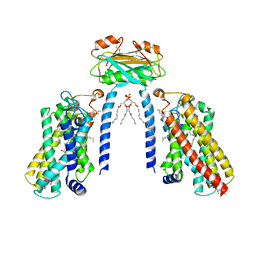 | | Cryo-EM structure of the C26-CoA-bound Lac1-Lip1 complex | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, Ceramide synthase subunit LIP1, ... | | Authors: | Xie, T, Fang, Q, Gong, X. | | Deposit date: | 2023-04-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure and mechanism of a eukaryotic ceramide synthase complex.
Embo J., 42, 2023
|
|
8IZF
 
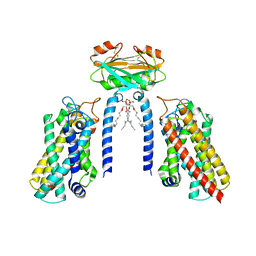 | | Cryo-EM structure of the Lac1-Lip1 (Lip1-S74F) complex | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, Ceramide synthase subunit LIP1 | | Authors: | Xie, T, Fang, Q, Gong, X. | | Deposit date: | 2023-04-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure and mechanism of a eukaryotic ceramide synthase complex.
Embo J., 42, 2023
|
|
7D0A
 
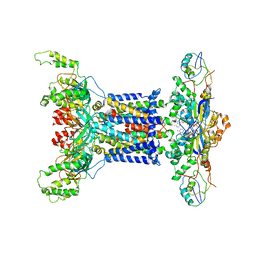 | | Acinetobacter MlaFEDB complex in ADP-vanadate trapped Vclose conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D09
 
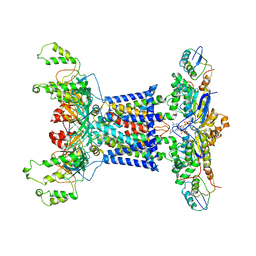 | | Acinetobacter MlaFEDB complex in ATP-bound Vtrans2 conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D08
 
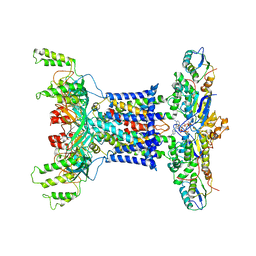 | | Acinetobacter MlaFEDB complex in ATP-bound Vtrans1 conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D06
 
 | | Cryo EM structure of the nucleotide free Acinetobacter MlaFEDB complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ABC transporter ATP-binding protein, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7CGN
 
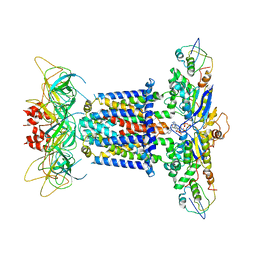 | | The overall structure of the MlaFEDB complex in ATP-bound EQtall conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7CGE
 
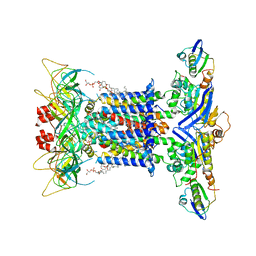 | | The overall structure of nucleotide free MlaFEDB complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7CH0
 
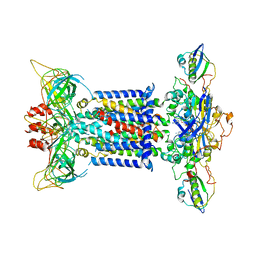 | | The overall structure of the MlaFEDB complex in ATP-bound EQclose conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
8GS9
 
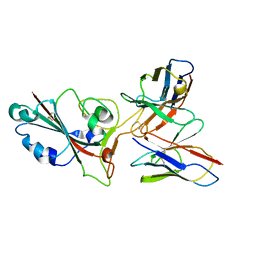 | | SARS-CoV-2 BA.2 spike RBD in complex bound with VacBB-551 | | Descriptor: | Heavy chain of VacBB-551, Light chain of VacBB-551, Spike glycoprotein | | Authors: | Liu, C.C, Ju, B, Shen, S.L, Zhang, Z. | | Deposit date: | 2022-09-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Omicron BQ.1.1 and XBB.1 unprecedentedly escape broadly neutralizing antibodies elicited by prototype vaccination.
Cell Rep, 42, 2023
|
|
8WCT
 
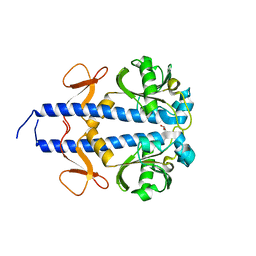 | |
8ZBQ
 
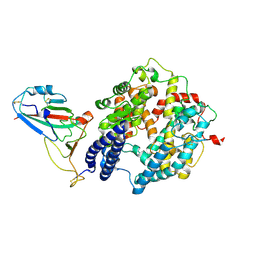 | | Local map of Omicron Subvariant JN.1 RBD with ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Yan, R.H, Yang, H.N. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-18 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants.
Nat Commun, 15, 2024
|
|
7WP2
 
 | | Cryo-EM structure of SARS-CoV-2 C.1.2 S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP0
 
 | | Cryo-EM structure of SARS-CoV-2 Delta S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c335_light_IGLV1-40_IGLJ3, Spike protein S1, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spikeprotein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
