8BQW
 
 | | Cryo-EM structure of alpha-synuclein filaments doublet from Juvenile-onset synucleinopathy | | Descriptor: | Alpha-synuclein, Unknown protein | | Authors: | Yang, Y, Garringer, J.H, Shi, Y, Lovestam, S, Peak-Chew, S.Y, Zhang, X.J, Kotecha, A, Bacioglu, M, Koto, A, Takao, M, Spillantini, G.M, Ghetti, B, Vidal, R, Murzin, G.A, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2023-01-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | New SNCA mutation and structures of alpha-synuclein filaments from juvenile-onset synucleinopathy.
Acta Neuropathol, 145, 2023
|
|
8BQV
 
 | | Cryo-EM structure of alpha-synuclein singlet filament from Juvenile-onset synucleinopathy | | Descriptor: | Alpha-synuclein, Unknown protein | | Authors: | Yang, Y, Garringer, J.H, Shi, Y, Lovestam, S, Peak-Chew, S.Y, Zhang, X.J, Kotecha, A, Bacioglu, M, Koto, A, Takao, M, Spillantini, G.M, Ghetti, B, Vidal, R, Murzin, G.A, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2023-01-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | New SNCA mutation and structures of alpha-synuclein filaments from juvenile-onset synucleinopathy.
Acta Neuropathol, 145, 2023
|
|
3KDU
 
 | |
3KDT
 
 | |
6T6B
 
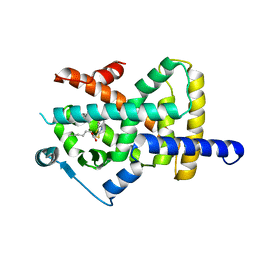 | | Crystal structure of PPARgamma in complex with compound 16 (MF27) | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Ni, X, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
8CRL
 
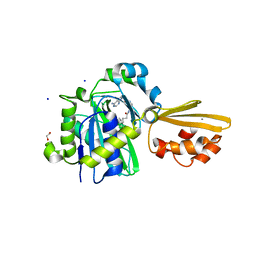 | | Crystal structure of LplA1 in complex with the inhibitor C3 (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CRI
 
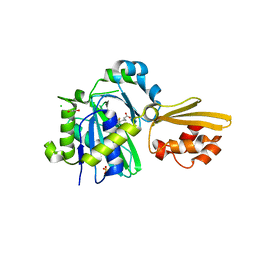 | | Crystal structure of LplA1 in complex with lipoic acid (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LIPOIC ACID, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CRJ
 
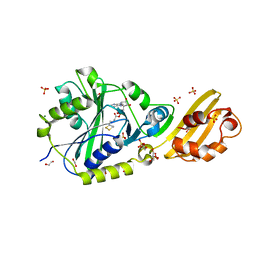 | | Crystal structure of LplA1 in complex with lipoyl-AMP (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, GLYCEROL, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5YJO
 
 | |
2RG1
 
 | | Crystal structure of E. coli WrbA apoprotein | | Descriptor: | CHLORIDE ION, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Lapkouski, M, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
6IJL
 
 | |
2R96
 
 | | Crystal structure of E. coli WrbA in complex with FMN | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
2R97
 
 | | Crystal structure of E. coli WrbA in complex with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
5X57
 
 | | Structure of GAR domain of ACF7 | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, NICKEL (II) ION | | Authors: | Yang, F, Wang, T, Zhang, Y, Wu, X.Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | ACF7 regulates inflammatory colitis and intestinal wound response by orchestrating tight junction dynamics.
Nat Commun, 8, 2017
|
|
4Z6G
 
 | | Structure of NT domain | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, PHOSPHATE ION | | Authors: | Yang, F, Zhang, Y. | | Deposit date: | 2015-04-05 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | In vivo epidermal migration requires focal adhesion targeting of ACF7.
Nat Commun, 7, 2016
|
|
1HG4
 
 | |
5PGU
 
 | |
5PGX
 
 | |
5PGY
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PGZ
 
 | | CRYSTAL STRUCTURE OF MURINE 11BETA- HYDROXYSTEROIDDEHYDROGENASE COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PGW
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3- HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE | | Descriptor: | 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PGV
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE | | Descriptor: | 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
7C2S
 
 | | Helical reconstruction of Dengue virus serotype 3 complexed with Fab C10 | | Descriptor: | Heavy chain of Fab C10, envelope protein, light chain of Fab C10 | | Authors: | Morrone, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Zhang, S, Lok, S.M. | | Deposit date: | 2020-05-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | High flavivirus structural plasticity demonstrated by a non-spherical morphological variant.
Nat Commun, 11, 2020
|
|
7C2T
 
 | | Helical reconstruction of Zika virus complexed with Fab C10 | | Descriptor: | Heavy chain from Fab C10, Light chain from Fab C10, M protein, ... | | Authors: | Morrone, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Zhang, S, Lok, S.M. | | Deposit date: | 2020-05-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | High flavivirus structural plasticity demonstrated by a non-spherical morphological variant.
Nat Commun, 11, 2020
|
|
1F4I
 
 | | SOLUTION STRUCTURE OF THE HHR23A UBA(2) MUTANT P333E, DEFICIENT IN BINDING THE HIV-1 ACCESSORY PROTEIN VPR | | Descriptor: | UV EXCISION REPAIR PROTEIN PROTEIN RAD23 HOMOLOG A | | Authors: | Withers-Ward, E.S, Mueller, T.D, Chen, I.S, Feigon, J. | | Deposit date: | 2000-06-07 | | Release date: | 2000-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr.
Biochemistry, 39, 2000
|
|
