3TMA
 
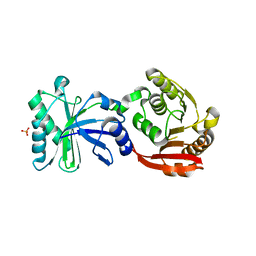 | | Crystal structure of TrmN from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, methyltransferase | | Authors: | Fislage, M, Roovers, M, Tuszynska, I, Bujnicki, J.M, Droogmans, L, Versees, W. | | Deposit date: | 2011-08-31 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life.
Nucleic Acids Res., 40, 2012
|
|
3TM4
 
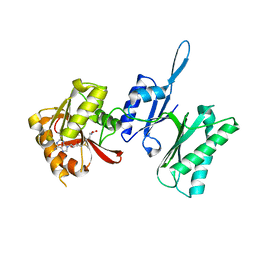 | | Crystal structure of Trm14 from Pyrococcus furiosus in complex with S-adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine N2-)-methyltransferase Trm14 | | Authors: | Fislage, M, Roovers, M, Tuszynska, I, Bujnicki, J.M, Droogmans, L, Versees, W. | | Deposit date: | 2011-08-31 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life.
Nucleic Acids Res., 40, 2012
|
|
2Y7H
 
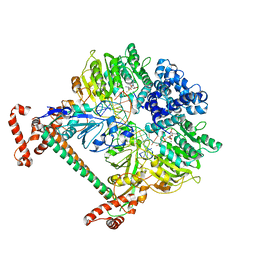 | | Atomic model of the DNA-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | 5'-D(*GP*TP*TP*CP*AP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*GP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*CP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*TP*GP*AP*AP*C)-3', S-ADENOSYLMETHIONINE, ... | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
2Y7C
 
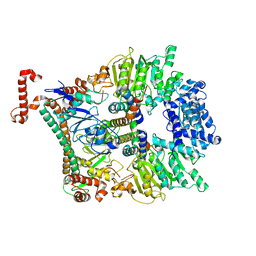 | | Atomic model of the Ocr-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | GENE 0.3 PROTEIN, TYPE I RESTRICTION ENZYME ECOKI M PROTEIN, TYPE-1 RESTRICTION ENZYME ECOKI SPECIFICITY PROTEIN | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
3HJ6
 
 | | Structure of Halothermothrix orenii fructokinase (FRK) | | Descriptor: | Fructokinase | | Authors: | Chua, T.K, Seetharaman, J, Kasprzak, J.M, Ng, C, Patel, B.K, Love, C, Bujnicki, J.M, Sivaraman, J. | | Deposit date: | 2009-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a fructokinase homolog from Halothermothrix orenii
J.Struct.Biol., 171, 2010
|
|
6FQ3
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with lin-29A 5'UTR 13mer RNA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*GP*GP*AP*GP*UP*CP*CP*AP*AP*CP*UP*CP*C)-3') | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6FPT
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains | | Descriptor: | E3 ubiquitin-protein ligase TRIM71 | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6FQL
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with mab-10 3'UTR 13mer RNA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*UP*GP*CP*AP*UP*UP*UP*AP*AP*UP*GP*CP*A)-3') | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6G6S
 
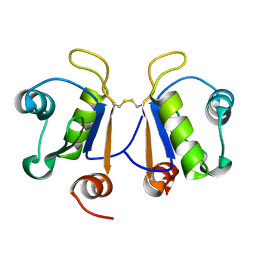 | | Crystal structure of human Acinus RNA recognition motif domain | | Descriptor: | Apoptotic chromatin condensation inducer in the nucleus | | Authors: | Fernandes, H, Czapinska, H, Grudziaz, K, Bujnicki, J.M, Nowacka, M. | | Deposit date: | 2018-04-03 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of human Acinus RNA recognition motif domain.
PeerJ, 6, 2018
|
|
3ISV
 
 | | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with acetate ion | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Skarina, T, Onopriyenko, O, Stam, J, Anderson, W.F, Savchenko, A, Bujnicki, J.M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with acetate ion
To be Published
|
|
3IST
 
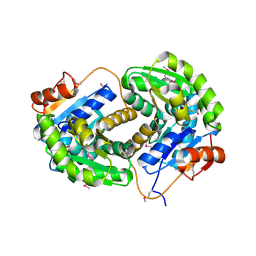 | | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid | | Descriptor: | CHLORIDE ION, Glutamate racemase, SUCCINIC ACID | | Authors: | Majorek, K.A, Chruszcz, M, Skarina, T, Onopriyenko, O, Stam, J, Anderson, W.F, Savchenko, A, Bujnicki, J.M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid
TO BE PUBLISHED
|
|
2PJD
 
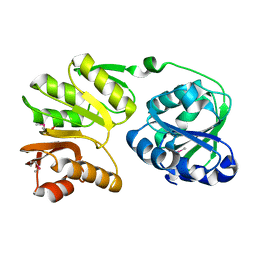 | | Crystal structure of 16S rRNA methyltransferase RsmC | | Descriptor: | Ribosomal RNA small subunit methyltransferase C | | Authors: | Sunita, S, Purta, E, Durawa, M, Tkaczuk, K.L, Bujnicki, J.M, Sivaraman, J. | | Deposit date: | 2007-04-16 | | Release date: | 2007-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional specialization of domains tandemly duplicated within 16S rRNA methyltransferase RsmC
Nucleic Acids Res., 35, 2007
|
|
7NYQ
 
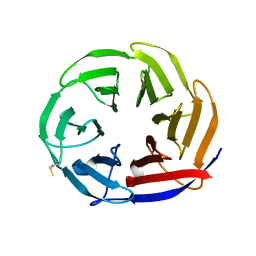 | | Crystal structure of the Mei-P26 NHL domain | | Descriptor: | Meiotic P26, isoform C | | Authors: | Salerno-Kochan, A, Gaik, M, Medenbach, J, Glatt, S. | | Deposit date: | 2021-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into RNA recognition and gene regulation by the TRIM-NHL protein Mei-P26.
Life Sci Alliance, 5, 2022
|
|
3C0K
 
 | |
3BM3
 
 | | Restriction endonuclease PspGI-substrate DNA complex | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*AP*TP*CP*CP*AP*GP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*GP*GP*AP*T)-3'), ... | | Authors: | Szczepanowski, R.H, Carpenter, M, Czapinska, H, Tamulaitis, G, Siksnys, V, Bhagwat, A, Bochtler, M. | | Deposit date: | 2007-12-12 | | Release date: | 2008-09-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A direct crystallographic demonstration that Type II restriction endonuclease PspGI flips nucleotides
To be Published
|
|
3LCV
 
 | | Crystal Structure of Antibiotic related Methyltransferase | | Descriptor: | S-ADENOSYLMETHIONINE, Sisomicin-gentamicin resistance methylase Sgm | | Authors: | Sivaraman, J, Husain, N. | | Deposit date: | 2010-01-11 | | Release date: | 2010-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the methylation of G1405 in 16S rRNA by aminoglycoside resistance methyltransferase Sgm from an antibiotic producer: a diversity of active sites in m7G methyltransferases.
Nucleic Acids Res., 2010
|
|
2R60
 
 | |
2R66
 
 | |
5A7Z
 
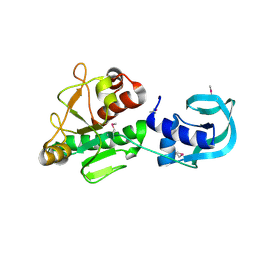 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.1 angstrom resolution. | | Descriptor: | TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
5A7Y
 
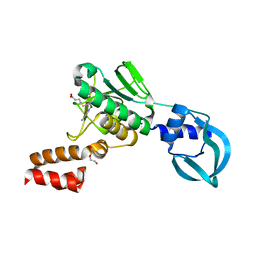 | | Crystal structure of Sulfolobus acidocaldarius Trm10 in complex with S-adenosylhomocysteine | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
5A7T
 
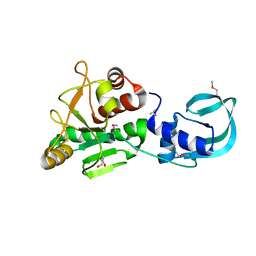 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.4 angstrom resolution. | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
7YR6
 
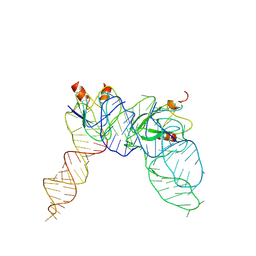 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers | | Descriptor: | RsmZ RNA, Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Ling, X, Yang, X, Wu, Y, Liu, T, Wei, X, Bujnick, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
2FCA
 
 | | The structure of BsTrmB | | Descriptor: | POTASSIUM ION, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Zegers, I, Van Vliet, F, Bujnicki, J, Kosinski, J, Gigot, D, Droogmans, L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase.
Nucleic Acids Res., 34, 2006
|
|
2R68
 
 | |
4QPT
 
 | | Structural Investigation of hnRNP L | | Descriptor: | Heterogenous nuclear ribonucleoprotein L, PHOSPHATE ION | | Authors: | Blatter, M, Allain, F.H.-T. | | Deposit date: | 2014-06-25 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | The Signature of the Five-Stranded vRRM Fold Defined by Functional, Structural and Computational Analysis of the hnRNP L Protein.
J.Mol.Biol., 427, 2015
|
|
