5IT5
 
 | | Thermus thermophilus PilB core ATPase region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP binding motif-containing protein PilF, MAGNESIUM ION, ... | | Authors: | Mancl, J, Robinson, H, Black, W, Yang, Z, Schubot, F. | | Deposit date: | 2016-03-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Crystal Structure of a Type IV Pilus Assembly ATPase: Insights into the Molecular Mechanism of PilB from Thermus thermophilus.
Structure, 24, 2016
|
|
3ZTH
 
 | | Crystal structure of Stu0660 of Streptococcus thermophilus | | Descriptor: | MAGNESIUM ION, STU0660 | | Authors: | Lee, K.-H, Robinson, H, Oh, B.-H. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification, Structural, and Biochemical Characterization of a Group of Large Csn2 Proteins Involved in Crispr-Mediated Bacterial Immunity.
Proteins, 80, 2012
|
|
1UII
 
 | |
4TYX
 
 | | Structure of aquoferric sperm whale myoglobin L29H/F33Y/F43H/S92A mutant | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhagi-Damodaran, A, Petrik, I.D, Robinson, H, Lu, Y. | | Deposit date: | 2014-07-09 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Systematic tuning of heme redox potentials and its effects on O2 reduction rates in a designed oxidase in myoglobin.
J.Am.Chem.Soc., 136, 2014
|
|
5TCB
 
 | |
6WJB
 
 | | UDP-GlcNAc C4-epimerase from Pseudomonas protegens in complex with NAD and UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Pfoh, R, Robinson, H, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
6WJ9
 
 | | UDP-GlcNAc C4-epimerase mutant S121A/Y146F from Pseudomonas protegens in complex with UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Willams, R.J, Whitney, J.C, Whitfield, G.B, Robinson, H, Parsek, M.R, Nitz, M, Harrison, J.J, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
5YDJ
 
 | | Crystal structure of anopheles gambiae acetylcholinesterase in complex with PMSF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Han, Q, Guan, H, Ding, H, Liao, C, Robinson, H, Li, J. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
5YAG
 
 | | Crystal structure of mosquito arylalkylamine N-Acetyltransferase like 5b/spermine N-Acetyltransferase | | Descriptor: | AAEL004827-PA, GLYCEROL | | Authors: | Han, Q, Guan, H, Robinson, H, Li, J. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of aaNAT5b as a spermine N-acetyltransferase in the mosquito, Aedes aegypti.
PLoS ONE, 13, 2018
|
|
5YDI
 
 | | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector anopheles gambiae, new crystal packing | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Han, Q, Guan, H, Ding, H, Liao, C, Robinson, H, Li, J. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
5YDH
 
 | | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector anopheles gambiae, 3.2 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Acetylcholinesterase, ... | | Authors: | Han, Q, Guan, H, Robinson, H, Ding, H, Liao, C, Li, J. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
5X61
 
 | | Crystal structure of Acetylcholinesterase Catalytic Subunit of the Malaria Vector Anopheles Gambiae, 3.4 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, ... | | Authors: | Han, Q, Robinson, H, Ding, H, Wong, D.M, Lam, P.C.H, Totrov, M.M, Carlier, P.R, Li, J. | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector Anopheles gambiae
Insect Sci., 2017
|
|
5EQ6
 
 | | Pseudomonas aeruginosa PilM bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EOY
 
 | | Pseudomonas aeruginosa SeMet-PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EOU
 
 | | Pseudomonas aeruginosa PilM:PilN1-12 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L, Howell, L.P. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5FC9
 
 | | Novel Purple Cupredoxin from Nitrosopumilus maritimus | | Descriptor: | Blue (Type 1) copper domain protein, COPPER (II) ION | | Authors: | Hosseinzadeh, P, Lu, Y, Robinson, H, Gao, Y.-G. | | Deposit date: | 2015-12-15 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Purple Cupredoxin from Nitrosopumilus maritimus Containing a Mononuclear Type 1 Copper Center with an Open Binding Site.
J.Am.Chem.Soc., 138, 2016
|
|
5EOX
 
 | | Pseudomonas aeruginosa PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
6BDC
 
 | |
4Z8Q
 
 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:AvrRxo1-ORF2 COMPLEX, SELENOMETHIONINE SUBSTITUTED. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4ZV4
 
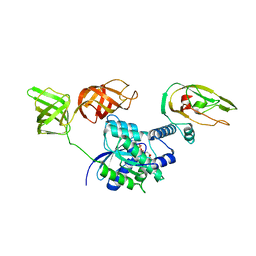 | | Structure of Tse6 in complex with EF-Tu | | Descriptor: | Elongation factor Tu, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Whitney, J.C, Sawai, S, Robinson, H, Mougous, J.D. | | Deposit date: | 2015-05-18 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.504 Å) | | Cite: | An Interbacterial NAD(P)(+) Glycohydrolase Toxin Requires Elongation Factor Tu for Delivery to Target Cells.
Cell, 163, 2015
|
|
4Z8T
 
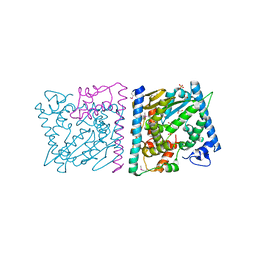 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:AvrRxo1-ORF2 WITH SULPHATE IONS | | Descriptor: | ACETATE ION, AvrRxo1-ORF1, AvrRxo1-ORF2, ... | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z8V
 
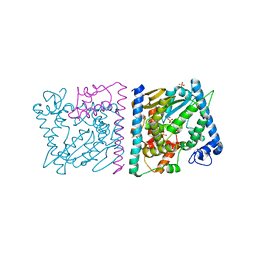 | | CRYSTAL STRUCTURE OF AVRRXO1-ORF1:-ORF2 COMPLEX, NATIVE. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4ZUY
 
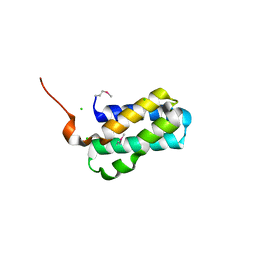 | | Structure of Tsi6 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Tsi6 | | Authors: | Whitney, J.C, Sawai, S, Robinson, H, Mougous, J.D. | | Deposit date: | 2015-05-18 | | Release date: | 2015-11-11 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | An Interbacterial NAD(P)(+) Glycohydrolase Toxin Requires Elongation Factor Tu for Delivery to Target Cells.
Cell, 163, 2015
|
|
4Z8U
 
 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:-ORF2 WITH ATP | | Descriptor: | ACETATE ION, AvrRxo1-ORF1, AvrRxo1-ORF2, ... | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4OJX
 
 | | crystal structure of yeast phosphodiesterase-1 in complex with GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3',5'-cyclic-nucleotide phosphodiesterase 1, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Tian, Y, Cui, W, Huang, M, Robinson, H, Wan, Y, Wang, Y, Ke, H. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Dual specificity and novel structural folding of yeast phosphodiesterase-1 for hydrolysis of second messengers cyclic adenosine and guanosine 3',5'-monophosphate.
Biochemistry, 53, 2014
|
|
