2GEC
 
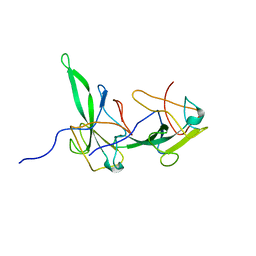 | | Structure of the N-terminal domain of avian infectious bronchitis virus nucleocapsid protein (strain Gray) in a novel dimeric arrangement | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-19 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
2GE7
 
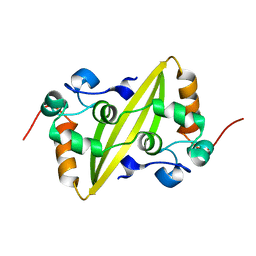 | | Structure of the C-terminal dimerization domain of infectious bronchitis virus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-18 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
2YI1
 
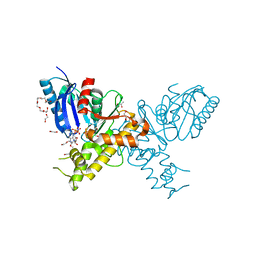 | | Crystal structure of N-Acetylmannosamine kinase in complex with N- acetyl mannosamine 6-phosphate and ADP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Reutter, W, Fan, H, Saenger, W, Moniot, S. | | Deposit date: | 2011-05-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
2YHW
 
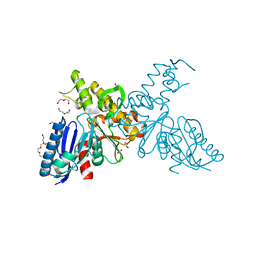 | | High-resolution crystal structures of N-Acetylmannosamine kinase: Insights about substrate specificity, activity and inhibitor modelling. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ACETATE ION, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Zimmer, R, Tauberger, E, Reutter, W, Saenger, W, Fan, H, Moniot, S. | | Deposit date: | 2011-05-08 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
2YHY
 
 | | Structure of N-Acetylmannosamine kinase in complex with N- acetylmannosamine and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Reutter, W, Fan, H, Saenger, W, Moniot, S. | | Deposit date: | 2011-05-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
6XZD
 
 | | Influenza C virus polymerase complex without chicken ANP32A - Subclass 2 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*GP*CP*CP*CP*UP*GP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-04 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6XZG
 
 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 3 | | Descriptor: | Influenza viral RNA (vRNA) promoter 47mer, LRRcap domain-containing protein, Polymerase acidic protein, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-04 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6XZR
 
 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 1 | | Descriptor: | LRRcap domain-containing protein, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6XZP
 
 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 4 | | Descriptor: | LRRcap domain-containing protein, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
2BGN
 
 | | HIV-1 Tat protein derived N-terminal nonapeptide Trp2-Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2005-01-03 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structures of HIV-1 Tat-Derived Nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) Bound to the Active Site of Dipeptidyl-Peptidase Iv (Cd26)
J.Biol.Chem., 280, 2005
|
|
2BGR
 
 | | Crystal structure of HIV-1 Tat derived nonapeptides Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2005-01-04 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of HIV-1 Tat-Derived Nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) Bound to the Active Site of Dipeptidyl-Peptidase Iv (Cd26)
J.Biol.Chem., 280, 2005
|
|
8R1L
 
 | | Structure of avian H5N1 influenza A polymerase in complex with human ANP32B. | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Staller, E, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-02 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of H5N1 influenza polymerase with ANP32B reveal mechanisms of genome replication and host adaptation.
Nat Commun, 15, 2024
|
|
8R1J
 
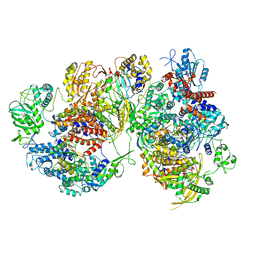 | | Structure of avian H5N1 influenza A polymerase dimer in complex with human ANP32B. | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Staller, E, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-02 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of H5N1 influenza polymerase with ANP32B reveal mechanisms of genome replication and host adaptation.
Nat Commun, 15, 2024
|
|
3KDH
 
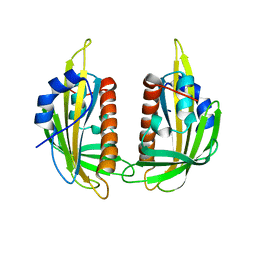 | | Structure of ligand-free PYL2 | | Descriptor: | Putative uncharacterized protein At2g26040 | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
3KDJ
 
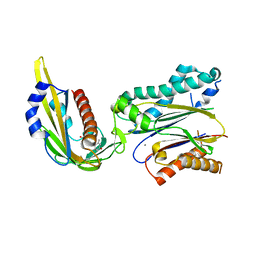 | | Complex structure of (+)-ABA-bound PYL1 and ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MANGANESE (II) ION, Protein phosphatase 2C 56, ... | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
3KDI
 
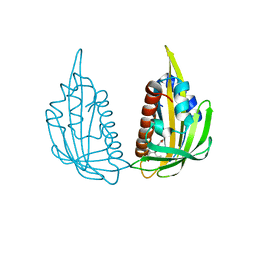 | | Structure of (+)-ABA bound PYL2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At2g26040 | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.379 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
4IC6
 
 | | Crystal structure of Deg8 | | Descriptor: | Protease Do-like 8, chloroplastic | | Authors: | Gong, W, Sun, W, Fan, H, Gao, F, Liu, L. | | Deposit date: | 2012-12-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IC5
 
 | | Crystal structure of Deg5 | | Descriptor: | CALCIUM ION, Protease Do-like 5, chloroplastic | | Authors: | Gong, W, Sun, W, Fan, H, Gao, F, Liu, L. | | Deposit date: | 2012-12-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7EPB
 
 | | Cryo-EM structure of LY354740-bound mGlu2 homodimer | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, Anti-RON nanobody, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPC
 
 | | Cryo-EM structure of inactive mGlu7 homodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPA
 
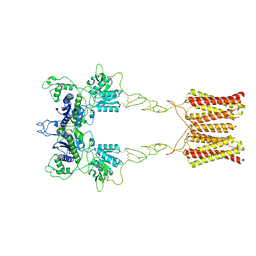 | | Cryo-EM structure of inactive mGlu2 homodimer | | Descriptor: | Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPD
 
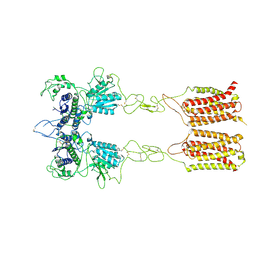 | | Cryo-EM structure of inactive mGlu2-7 heterodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7, Metabotropic glutamate receptor 2,Peptidylprolyl isomerase | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7FHB
 
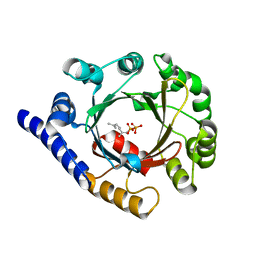 | | Structure of prenyltransferase from Streptomyces sp. (strain CL190) with bound GPP | | Descriptor: | GERANYL DIPHOSPHATE, Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHC
 
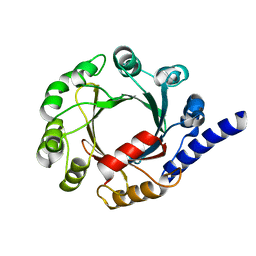 | | Structure of prenyltransferase mutant V49W from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHF
 
 | | Structure of prenyltransferase mutant V49W/Y288F/Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
