2D47
 
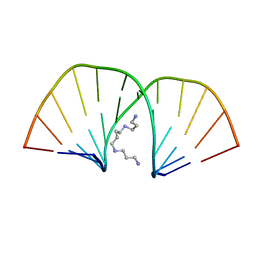 | | MOLECULAR STRUCTURE OF A COMPLETE TURN OF A-DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*GP*CP*GP*GP*GP*GP*G)-3'), SPERMINE | | Authors: | Verdaguer, N, Aymami, J, Fernandez-Forner, D, Fita, I, Coll, M, Huynh-Dinh, T, Igolen, J, Subirana, J.A. | | Deposit date: | 1991-10-02 | | Release date: | 1991-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of a complete turn of A-DNA.
J.Mol.Biol., 221, 1991
|
|
2D5V
 
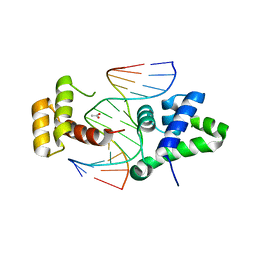 | | Crystal structure of HNF-6alpha DNA-binding domain in complex with the TTR promoter | | Descriptor: | 5'-D(*AP*TP*TP*AP*TP*TP*GP*AP*CP*TP*TP*AP*GP*A)-3', 5'-D(*TP*CP*TP*AP*AP*GP*TP*CP*AP*AP*TP*AP*AP*T)-3', ACETATE ION, ... | | Authors: | Iyaguchi, D, Yao, M, Watanabe, N, Nishihira, J, Tanaka, I. | | Deposit date: | 2005-11-07 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA recognition mechanism of the ONECUT homeodomain of transcription factor HNF-6
Structure, 15, 2007
|
|
6NES
 
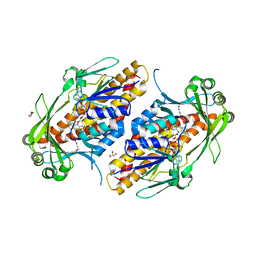 | | FAD-dependent monooxygenase TropB from T. stipitatus | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
3KRV
 
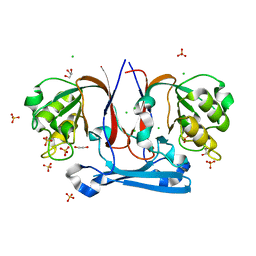 | | The Structure Of Potential Metal-Dependent Hydrolase With Cyclase Activity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rakonjac, N, Rezacova, P, Borek, D, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-19 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure Of Potential Metal-Dependent Hydrolase With Cyclase Activity
To be Published
|
|
2DKC
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the substrate complex | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
6NUK
 
 | | De novo designed protein Ferredog-Diesel | | Descriptor: | Ferredog-Diesel | | Authors: | Koepnick, B, Bick, M.J, DiMaio, F, Norgard-Solano, T, Baker, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
2DC6
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA073 complex | | Descriptor: | BENZYL N-({(2S,3S)-3-[(PROPYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-28 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
6NBH
 
 | | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein | | Descriptor: | CHOLESTEROL, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, L.-H, Ma, S, Sutkeviciute, I, Shen, D.-D, Zhou, X.E, de Waal, P.P, Li, C.-Y, Kang, Y, Clark, L.J, Jean-Alphonse, F.G, White, A.D, Xiao, K, Yang, D, Jiang, Y, Watanabe, T, Gardella, T.J, Melcher, K, Wang, M.-W, Vilardaga, J.-P, Xu, H.E, Zhang, Y. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and dynamics of the active human parathyroid hormone receptor-1.
Science, 364, 2019
|
|
2DC7
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA042 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-THREONYL-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCC
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA077 complex | | Descriptor: | BENZYL N-({(2S,3S)-3-[(BENZYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, CATHEPSIN B, GLYCEROL, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCD
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA078 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-({(2S,3S)-3-[(BENZYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
6NET
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus substrate complex | | Descriptor: | 2,4-dihydroxy-3,6-dimethylbenzaldehyde, CHLORIDE ION, FAD-dependent monooxygenase tropB, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6JD0
 
 | | Structure of mutant human cathepsin L, engineered for GAG binding | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cathepsin L1, ... | | Authors: | Choudhury, D, Biswas, S. | | Deposit date: | 2019-01-30 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure-guided protein engineering of human cathepsin L for efficient collagenolytic activity.
Protein Eng.Des.Sel., 34, 2021
|
|
2DCB
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA076 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCYL-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCA
 
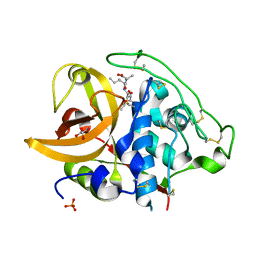 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA075 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCYL-L-ALANINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC9
 
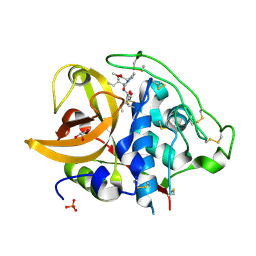 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA074Me complex | | Descriptor: | CATHEPSIN B, GLYCEROL, METHYL N-({(2S,3S)-3-[(PROPYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC8
 
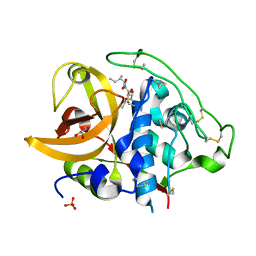 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA059 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DWN
 
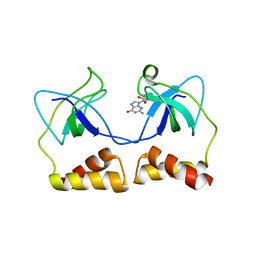 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | DNA (5'-D(*A*G)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
6NJH
 
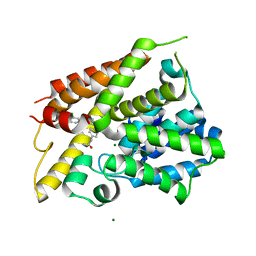 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48 | | Descriptor: | 2-(4-{[4-(3-chlorophenyl)-6-ethyl-1,3,5-triazin-2-yl]amino}phenyl)acetamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2019-01-03 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders.
J.Med.Chem., 62, 2019
|
|
7ZFR
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide bound in the reverse direction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DP alpha chain (DPA1*02:01), MHC class II HLA-DP beta chain (DPB1*01:01), ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
6Q2W
 
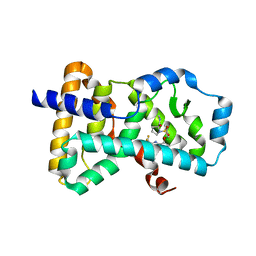 | | Crystal structure of human ROR gamma LBD in complex with a quinoline sulfonamide inverse agonist | | Descriptor: | (2~{S})-1-[2,4-bis(chloranyl)-3-[[4-imidazol-1-yl-2-(trifluoromethyl)quinolin-8-yl]oxymethyl]phenyl]sulfonyl-~{N}-methyl-pyrrolidine-2-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Ciesielski, F, Amaudrut, J, Argiriadi, M.A, Barth, M, Breinlinger, E.C, Calderwood, D.J, Cusack, K.P, Kort, M.E, Montalbetti, C, Potin, D, Poupardin, O, Spitzer, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of novel quinoline sulphonamide derivatives as potent, selective and orally active ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
2DH1
 
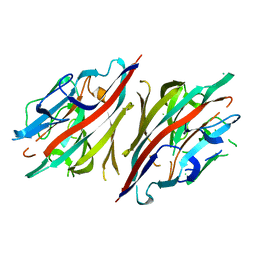 | | Crystal structure of peanut lectin lactose-azobenzene-4,4'-dicarboxylic acid-lactose complex | | Descriptor: | Galactose-binding lectin | | Authors: | Natchiar, S.K, Srinivas, O, Nivedita, M, Sagarika, D, Jayaraman, N, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.65 Å) | | Cite: | Multivalency in lectins - A crystallographic, modelling and light-scattering study involving peanut lectin and a bivalent ligand
Curr.Sci., 90, 2006
|
|
6NIN
 
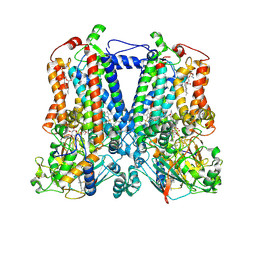 | | Rhodobacter sphaeroides bc1 with STIGMATELLIN A | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Cytochrome b, Cytochrome c1, ... | | Authors: | Xia, D, Zhou, F, Esser, L. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of bacterial cytochromebc1in complex with azoxystrobin reveals a conformational switch of the Rieske iron-sulfur protein subunit.
J.Biol.Chem., 294, 2019
|
|
2DUJ
 
 | | Crystal structure of the complex formed between proteinase K and a synthetic peptide Leu-Leu-Phe-Asn-Asp at 1.67 A resolution | | Descriptor: | CALCIUM ION, LLFND, NITRATE ION, ... | | Authors: | Singh, A.K, Singh, N, Somvanshi, R.K, Gupta, D, Sharma, S, Singh, T.P. | | Deposit date: | 2006-07-23 | | Release date: | 2006-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the complex of proteinase K with a specific lactoferrin peptide Val-Leu-Leu-His at 1.93 A resolution
To be Published
|
|
6NSJ
 
 | | CryoEM structure of Helicobacter pylori urea channel in closed state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | Authors: | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
