6DKH
 
 | | The crystal structure of L-idonate 5-dehydrogenase from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | L-idonate 5-dehydrogenase (NAD(P)(+)), ZINC ION | | Authors: | Tan, K, Evdokimova, E, McChesney, C, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | The crystal structure of L-idonate 5-dehydrogenase from Escherichia coli str. K-12 substr. MG1655
To Be Published
|
|
1RLH
 
 | | Structure of a conserved protein from Thermoplasma acidophilum | | Descriptor: | SODIUM ION, conserved hypothetical protein | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a conserved protein from T. acidophilum
To be Published
|
|
3MU0
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU8
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU4
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. First step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1EG2
 
 | | CRYSTAL STRUCTURE OF RHODOBACTER SPHEROIDES (N6 ADENOSINE) METHYLTRANSFERASE (M.RSRI) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MODIFICATION METHYLASE RSRI | | Authors: | Scavetta, R.D, Thomas, C.B, Walsh, M.A, Szegedi, S, Joachimiak, A, Gumport, R.I, Churchill, M.E.A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-02-11 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases.
Nucleic Acids Res., 28, 2000
|
|
6DZG
 
 | |
1G60
 
 | | Crystal Structure of Methyltransferase MboIIa (Moraxella bovis) | | Descriptor: | Adenine-specific Methyltransferase MboIIA, S-ADENOSYLMETHIONINE, SODIUM ION | | Authors: | Osipiuk, J, Walsh, M.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-11-02 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of MboIIA methyltransferase.
Nucleic Acids Res., 31, 2003
|
|
3U4G
 
 | | The Structure of CobT from Pyrococcus horikoshii | | Descriptor: | ACETIC ACID, NaMN:DMB phosphoribosyltransferase, SULFATE ION | | Authors: | Cuff, M.E, Evdokimova, E, Mursleen, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-07 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of CobT from Pyrococcus horikoshii
TO BE PUBLISHED
|
|
2QLT
 
 | | Crystal structure of an isoform of DL-glycerol-3-phosphatase, Rhr2p, from Saccharomyces cerevisiae | | Descriptor: | (DL)-glycerol-3-phosphatase 1, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of an isoform of DL-glycerol-3-phosphatase, Rhr2p from Saccharomyces cerevisiae.
To be Published
|
|
1TU1
 
 | | Crystal Structure of Protein of Unknown Function PA94 from Pseudomonas aeruginosa, Putative Regulator | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of hypothetical protein PA94 from Pseudomonas aeruginosa
To be Published
|
|
1TUA
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function APE0754 from Aeropyrum pernix | | Descriptor: | Hypothetical protein APE0754 | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein APE0754 from Aeropyrum pernix
To be Published
|
|
1TP6
 
 | | 1.5 A Crystal Structure of a NTF-2 Like Protein of Unknown Function PA1314 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA1314 | | Authors: | Zhang, R, Xu, L.X, savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA1314 from Pseudomonas aeruginosa
To be Published
|
|
2QIB
 
 | | Crystal structure of tetR-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | HEXAETHYLENE GLYCOL, tetR-family transcriptional regulator | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-03 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of tetR-family transcriptional regulator from Streptomyces coelicolor.
To be Published
|
|
2R8W
 
 | | The crystal structure of dihydrodipicolinate synthase (Atu0899) from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETATE ION, AGR_C_1641p, CHLORIDE ION | | Authors: | Tan, K, Dong, A, Xu, X, Gu, J, Zheng, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of dihydrodipicolinate synthase (Atu0899) from Agrobacterium tumefaciens str. C58.
To be Published
|
|
1SQU
 
 | |
1DNO
 
 | | A-DNA/RNA DODECAMER R(GCG)D(TATACGC) MG BINDING SITES | | Descriptor: | DNA/RNA (5'-R(*GP*CP*GP)-D(*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-16 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
1U60
 
 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
1DCR
 
 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G-A BASE PAIRS | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*(BRU)P*GP*AP*GP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-11-05 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
1D9R
 
 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G-A BASE PAIRS | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*(BRU)P*GP*AP*GP*G)-3', COBALT HEXAMMINE(III) | | Authors: | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-10-29 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
6X1L
 
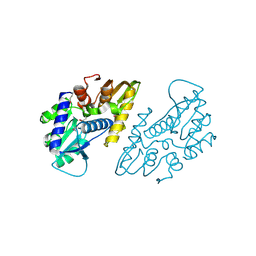 | | The crystal structure of a functional uncharacterized protein KP1_0663 from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | | Descriptor: | WbbZ protein | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
2QKO
 
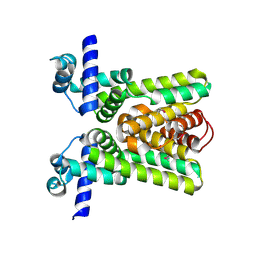 | | Crystal structure of transcriptional regulator RHA06399 from Rhodococcus sp. RHA1 | | Descriptor: | Possible transcriptional regulator, TetR family protein | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-11 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of tetR family protein.
To be Published
|
|
6WGP
 
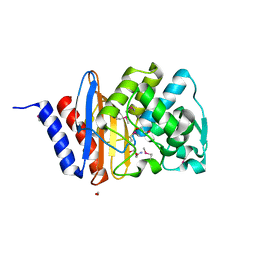 | | The crystal structure of a beta lactamase from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a beta lactamase from Xanthomonas campestris pv. campestris str. ATCC 33913
To Be Published
|
|
2QMA
 
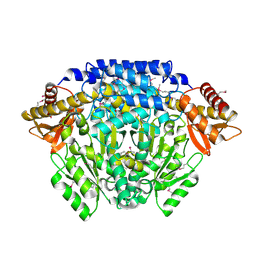 | | Crystal structure of glutamate decarboxylase domain of diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, Diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase | | Authors: | Osipiuk, J, Sather, A, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystal structure of glutamate decarboxylase domain of diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase from Vibrio parahaemolyticus.
To be Published
|
|
6WRH
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Welk, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
