2MJV
 
 | |
7WIN
 
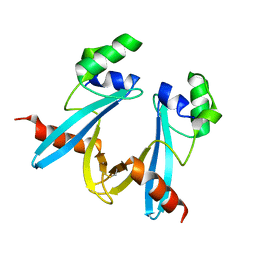 | |
7VON
 
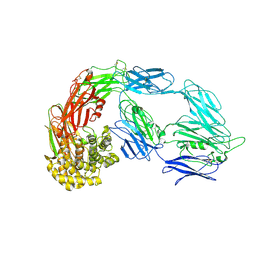 | | Native alpha-2-macroglobulin monomer | | Descriptor: | Alpha-2-macroglobulin | | Authors: | Huang, X, Wang, Y. | | Deposit date: | 2021-10-14 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structures reveal the dynamic transformation of human alpha-2-macroglobulin working as a protease inhibitor.
Sci China Life Sci, 65, 2022
|
|
7VOO
 
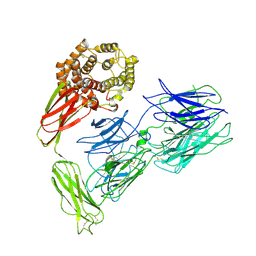 | | Induced alpha-2-macroglobulin monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Huang, X, Wang, Y, Ping, Z. | | Deposit date: | 2021-10-14 | | Release date: | 2022-10-19 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal the dynamic transformation of human alpha-2-macroglobulin working as a protease inhibitor.
Sci China Life Sci, 65, 2022
|
|
2L84
 
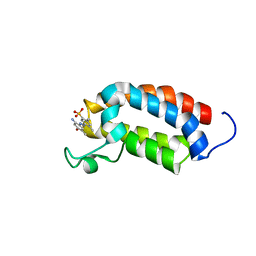 | | Solution NMR structures of CBP bromodomain with small molecule j28 | | Descriptor: | 5-[(E)-(2-amino-4-hydroxy-5-methylphenyl)diazenyl]-2,4-dimethylbenzenesulfonic acid, CREB-binding protein | | Authors: | Borah, J.C, Mujtaba, S, Karakikes, I, Zeng, L, Muller, M, Patel, J, Moshkina, N, Morohashi, K, Zhang, W, Gerona-Navarro, G, Hajjar, R.J, Zhou, M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Binding to the Coactivator CREB-Binding Protein Blocks Apoptosis in Cardiomyocytes.
Chem.Biol., 18, 2011
|
|
4B2T
 
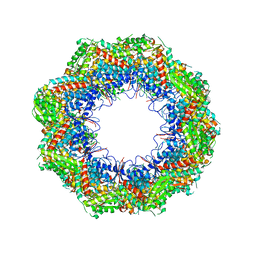 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT ALPHA, T-COMPLEX PROTEIN 1 SUBUNIT BETA, T-COMPLEX PROTEIN 1 SUBUNIT DELTA, ... | | Authors: | Kalisman, N, Schroeder, G.F, Levitt, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-03-20 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
2L85
 
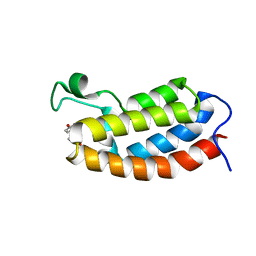 | | Solution NMR structures of CBP bromodomain with small molecule of HBS | | Descriptor: | 4-[(E)-(4-hydroxyphenyl)diazenyl]benzenesulfonic acid, CREB-binding protein | | Authors: | Borah, J.C, Mujtaba, S, Karakikes, I, Zeng, L, Muller, M, Patel, J, Moshkina, N, Morohashi, K, Zhang, W, Gerona-Navarro, G, Hajjar, R.J, Zhou, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Binding to the Coactivator CREB-Binding Protein Blocks Apoptosis in Cardiomyocytes.
Chem.Biol., 18, 2011
|
|
5T0Q
 
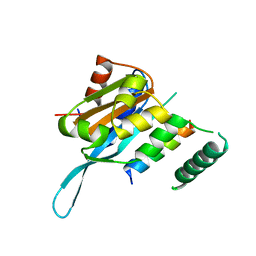 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 Jas domain [166-192] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T0F
 
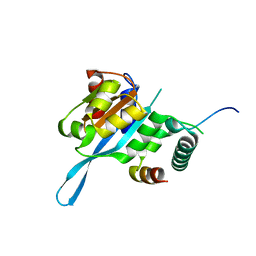 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 CMID domain [16-58] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2X12
 
 | |
3C8G
 
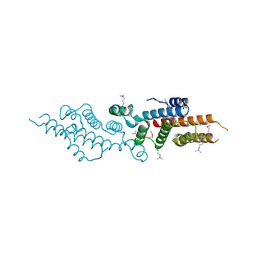 | | Crystal structure of a possible transciptional regulator YggD from Shigella flexneri 2a str. 2457T | | Descriptor: | ACETATE ION, Putative transcriptional regulator | | Authors: | Tan, K, Borovilos, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
7FEF
 
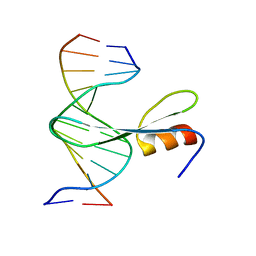 | | Crystal structure of AtMBD6 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain-containing protein 6 | | Authors: | Wu, Z.B, Liu, K, Min, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
2LLJ
 
 | | Structure of a bis-naphthalene bound to a thymine-thymine DNA mismatch | | Descriptor: | 6,22-dioxa-3,9,19,25-tetraazoniapentacyclo[25.5.3.3~11,17~.0~14,37~.0~30,34~]octatriaconta-1(33),11(38),12,14(37),15,17(36),27,29,31,34-decaene, DNA (5'-D(*CP*GP*TP*CP*GP*TP*AP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*TP*TP*CP*GP*AP*CP*G)-3') | | Authors: | Jourdan, M, Granzhan, A, Guillot, R, Dumy, P, Teulade-Fichou, M. | | Deposit date: | 2011-11-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Double threading through DNA: NMR structural study of a bis-naphthalene macrocycle bound to a thymine-thymine mismatch.
Nucleic Acids Res., 40, 2012
|
|
2LL9
 
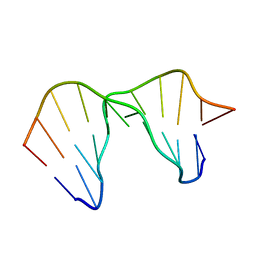 | | Solution structure of a DNA containing a thymime-thymine mismatch | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*GP*TP*AP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*TP*TP*CP*GP*AP*CP*G)-3') | | Authors: | Jourdan, M, Granzhan, A, Guillot, R, Dumy, P, Teulade-Fichou, M. | | Deposit date: | 2011-11-03 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Double threading through DNA: NMR structural study of a bis-naphthalene macrocycle bound to a thymine-thymine mismatch.
Nucleic Acids Res., 40, 2012
|
|
2MFQ
 
 | |
5C94
 
 | | Infectious bronchitis virus nsp9 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-structural protein 9 | | Authors: | Chen, C, Dou, Y, Yang, H, Su, D. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9.
Protein Sci., 26, 2017
|
|
4Z5V
 
 | |
4PHR
 
 | | Domain of unknown function 1792 (DUF1792) with manganese | | Descriptor: | ACETATE ION, MANGANESE (II) ION, Putative glycosyltransferase (GalT1), ... | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold.
Nat Commun, 5, 2014
|
|
6WE0
 
 | | Wheat dwarf virus Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*AP*CP*C)-3'), MANGANESE (II) ION, Replication-associated protein | | Authors: | Tompkins, K, Litzau, L.A, Shi, K, Nelson, A, Evans III, R.L, Gordon, W.R. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
4PFX
 
 | |
2KUB
 
 | | Solution structure of the alpha subdomain of the major non-repeat unit of Fap1 fimbriae of Streptococcus parasanguis | | Descriptor: | Fimbriae-associated protein Fap1 | | Authors: | Ramboarina, S, Garnett, J.A, Bodey, A, Simpson, P, Bardiaux, B, Nilges, M, Matthews, S. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into serine-rich fimbriae from gram-positive bacteria.
J.Biol.Chem., 2010
|
|
4PHS
 
 | |
2LMZ
 
 | |
4L1X
 
 | | Crystal Structuer of Human 3-alpha Hydroxysteroid Dehydrogenase Type 3 V54L Mutant in Complex with NADP+ and Progesterone | | Descriptor: | Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human 3-alpha hydroxysteroid dehydrogenase type 3 (3 alpha-HSD3): The V54L mutation restricting the steroid alternative binding and enhancing the 20 alpha-HSD activity
J.Steroid Biochem.Mol.Biol., 141, 2014
|
|
3DWH
 
 | |
