7CWM
 
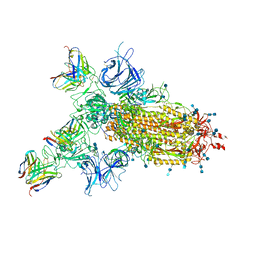 | |
7CWN
 
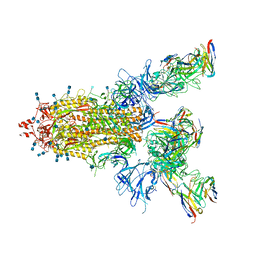 | | P17-H014 Fab cocktail in complex with SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, N, Wang, X. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
6M2B
 
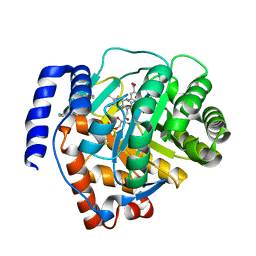 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with S416 | | Descriptor: | 2-[(E)-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]-methyl-hydrazinylidene]methyl]benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Li, H. | | Deposit date: | 2020-02-27 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2.
Protein Cell, 11, 2020
|
|
3G6L
 
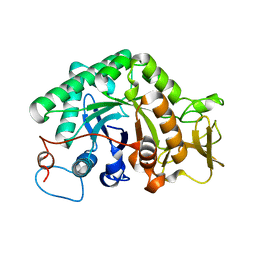 | |
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | Descriptor: | CAFFEINE, Chitinase | | Authors: | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | Deposit date: | 2009-02-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
7CWO
 
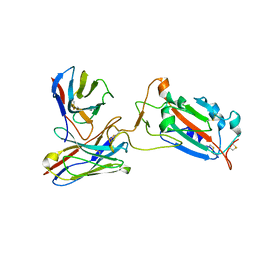 | | SARS-CoV-2 spike protein RBD and P17 fab complex | | Descriptor: | Spike glycoprotein, heavy chain of P17 Fab, light chain of P17 Fab | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
6M1I
 
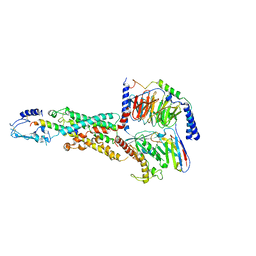 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
3NX2
 
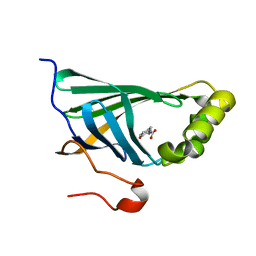 | | Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase in complex with substrate analogues | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
4LWI
 
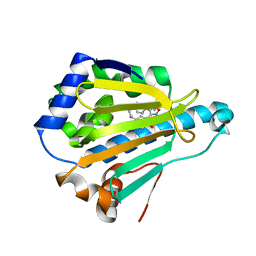 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWF
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ3 | | Descriptor: | 4-(5-amino-1,2-oxazol-3-yl)-6-(propan-2-yl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWG
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ4 | | Descriptor: | 1-(5-chloro-2,4-dihydroxyphenyl)-2-(4-methoxyphenyl)ethanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
9H8E
 
 | | Crystal structure of HPK1 T165E/S171E in complex with compound 13 | | Descriptor: | 5-(methylamino)-6-(3-methylimidazo[4,5-c]pyridin-7-yl)-3-[(4-morpholin-4-ylphenyl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Schimpl, M, Pflug, A. | | Deposit date: | 2024-10-29 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Discovery and Optimization of Pyrazine Carboxamide AZ3246, a Selective HPK1 Inhibitor.
J.Med.Chem., 68, 2025
|
|
9H8D
 
 | | Crystal structure of HPK1 T165E/S171E in complex with compound 6 | | Descriptor: | 6-(1-methylbenzimidazol-4-yl)-3-[(4-morpholin-4-ylphenyl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Schimpl, M, Pflug, A. | | Deposit date: | 2024-10-29 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Discovery and Optimization of Pyrazine Carboxamide AZ3246, a Selective HPK1 Inhibitor.
J.Med.Chem., 68, 2025
|
|
9H8F
 
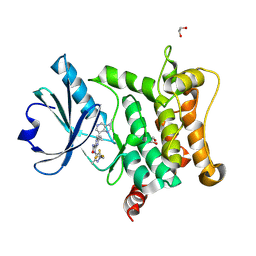 | | Crystal structure of HPK1 T165E/S171E in complex with pyrazine carboxamide inhibitor AZ3246 (compound 24) | | Descriptor: | 1,2-ETHANEDIOL, 5-cyclopropyl-6-(3-methylimidazo[4,5-c]pyridin-7-yl)-3-[[3-methyl-1-[2,2,2-tris(fluoranyl)ethyl]pyrazol-4-yl]amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Schimpl, M, Pflug, A. | | Deposit date: | 2024-10-29 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Discovery and Optimization of Pyrazine Carboxamide AZ3246, a Selective HPK1 Inhibitor.
J.Med.Chem., 68, 2025
|
|
4LWH
 
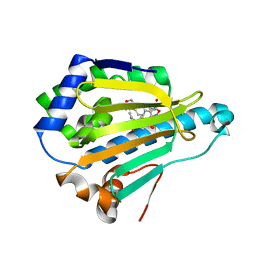 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7DA4
 
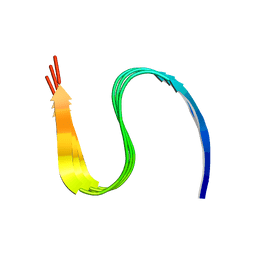 | | Cryo-EM structure of amyloid fibril formed by human RIPK3 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Zhao, K, Ma, Y.Y, Sun, Y.P, Li, D, Liu, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4EQ3
 
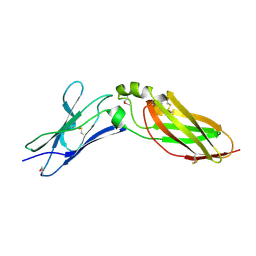 | | Crystal Structure Analysis of Selenomethionine (Se-Met) Substituted Chicken Interferon Gamma Receptor Alpha Chain | | Descriptor: | Interferon gamma receptor 1 | | Authors: | Ping, Z, Qi, J, Lu, G, Shi, Y, Wang, X, Gao, G.F, Wang, M. | | Deposit date: | 2012-04-18 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of the interferon gamma receptor alpha chain from chicken reveals an undetected extra helix compared with the human counterparts.
J.Interferon Cytokine Res., 34, 2014
|
|
4LWE
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ2 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-(5-chloro-2,4-dihydroxyphenyl)-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]acetamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4EQ2
 
 | | Crystal Structure Analysis of Chicken Interferon Gamma Receptor Alpha Chain | | Descriptor: | Interferon gamma receptor 1 | | Authors: | Ping, Z, Qi, J, Lu, G, Shi, Y, Wang, X, Gao, G.F, Wang, M. | | Deposit date: | 2012-04-18 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structure of the interferon gamma receptor alpha chain from chicken reveals an undetected extra helix compared with the human counterparts.
J.Interferon Cytokine Res., 34, 2014
|
|
7XEY
 
 | | EDS1-PAD4 complexed with pRib-ADP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-beta-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Huang, S, Jia, A, Xiao, Y. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Science, 377, 2022
|
|
2N7L
 
 | |
