1LXJ
 
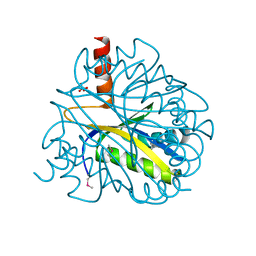 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | Descriptor: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
4ZMH
 
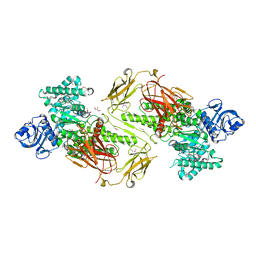 | | Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Nocek, B, Cui, H, Wang, W, Savchenko, A. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Biochemical and Structural Characterization of a Five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T.
J.Biol.Chem., 291, 2016
|
|
4QN8
 
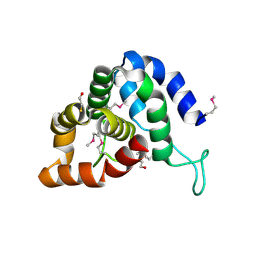 | | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | BETA-MERCAPTOETHANOL, VipE | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
1NR9
 
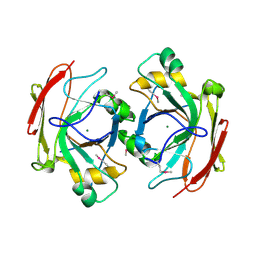 | | Crystal Structure of Escherichia coli 1262 (APC5008), Putative Isomerase | | Descriptor: | MAGNESIUM ION, Protein YCGM | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-24 | | Release date: | 2003-07-29 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Escherichia coli Putative Isomerase EC1262 (APC5008)
To be Published
|
|
5JD8
 
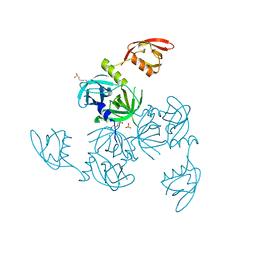 | | Crystal structure of the serine endoprotease from Yersinia pestis | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DI(HYDROXYETHYL)ETHER, Periplasmic serine peptidase DegS, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the serine endoprotease from Yersinia pestis
To Be Published
|
|
1NQK
 
 | | Structural Genomics, Crystal structure of Alkanesulfonate monooxygenase | | Descriptor: | Alkanesulfonate monooxygenase | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the protein
Alkanesulfonate monooxygenase from E. Coli
To be Published
|
|
1R0U
 
 | | Crystal structure of ywiB protein from Bacillus subtilis | | Descriptor: | GLYCEROL, protein ywiB | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of ywiB protein from Bacillus subtilis
to be published
|
|
4RXI
 
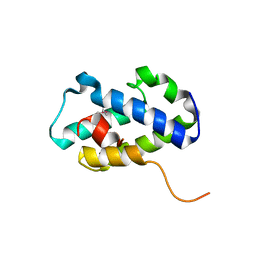 | | Structure of C-terminal domain of uncharacterized protein from Legionella pneumophila | | Descriptor: | hypothetical protein lpg0944 | | Authors: | Cuff, M, Nocek, B, Evdokimova, E, Egorova, O, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-11 | | Release date: | 2015-05-06 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
6MGL
 
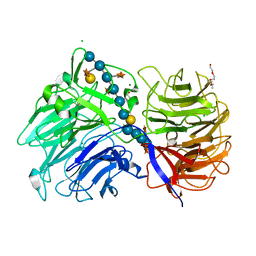 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, D60A mutant in complex with XXLG and XGXXLG xyloglucan | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Watanabe, N, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
4O2I
 
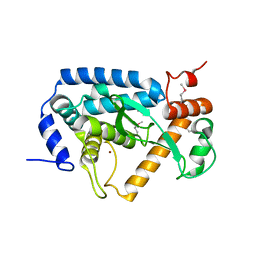 | | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium | | Descriptor: | Non-LEE encoded type III effector C, ZINC ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium
To be Published
|
|
4NE4
 
 | | Crystal structure of ABC transporter substrate binding protein ProX from Agrobacterium tumefaciens cocrystalized with BTB | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, substrate binding protein (Proline/glycine/betaine), ... | | Authors: | Tkaczuk, K.L, Nicholls, R, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of ABC transporter substrate binding protein ProX from Agrobacterium tumefaciens cocrystalized with BTB
To be Published
|
|
1Q8B
 
 | |
5IQJ
 
 | | 1.9 Angstrom Crystal Structure of Protein with Unknown Function from Vibrio cholerae. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Skarina, T, Seed, K.D, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Protein with Unknown Function from Vibrio cholerae.
To Be Published
|
|
6MGJ
 
 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
6MGK
 
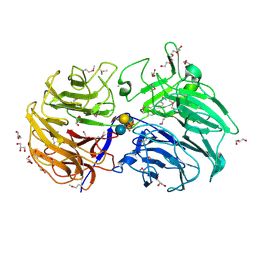 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
5KKO
 
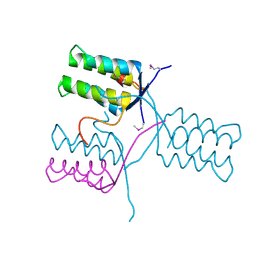 | | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein | | Descriptor: | Uncharacterised protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-22 | | Release date: | 2016-09-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein
To Be Published
|
|
5KVR
 
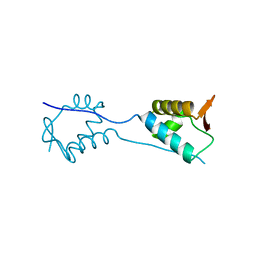 | | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073 | | Descriptor: | Pyruvate dehydrogenase complex repressor | | Authors: | Brunzelle, J.S, Wawrzak, Z, Sandoval, J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073
To Be Published
|
|
1RLH
 
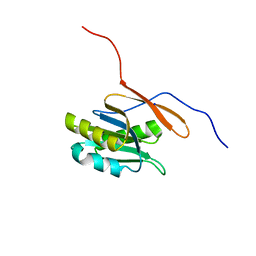 | | Structure of a conserved protein from Thermoplasma acidophilum | | Descriptor: | SODIUM ION, conserved hypothetical protein | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a conserved protein from T. acidophilum
To be Published
|
|
4R0J
 
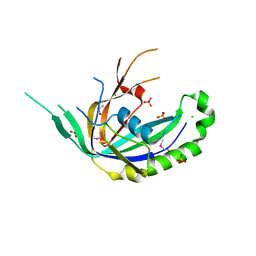 | | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans
To be Published
|
|
1TJN
 
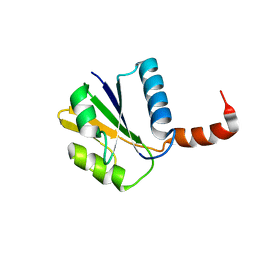 | | Crystal structure of hypothetical protein af0721 from Archaeoglobus fulgidus | | Descriptor: | Sirohydrochlorin cobaltochelatase | | Authors: | Yin, J, Xu, X.L, Cuff, M, Walker, J.R, Edwards, A, Savchenko, A, James, M.N.G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of af0721: a hypothetical protein bearing sequence similarity with class II chelatases in cobalamin synthesis
To be Published
|
|
4RD8
 
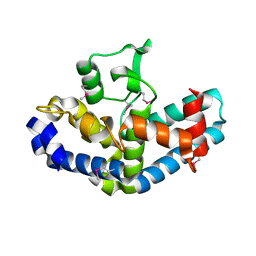 | | The crystal structure of a functionally-unknown protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of a functionally-unknown protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
1KS2
 
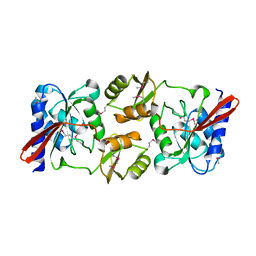 | | Crystal Structure Analysis of the rpiA, Structural Genomics, protein EC1268. | | Descriptor: | protein EC1268, RPIA | | Authors: | Zhang, R, Joachimiak, A, Edwards, A.M, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-10 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle.
STRUCTURE, 11, 2003
|
|
4PSU
 
 | | Crystal structure of alpha/beta hydrolase from Rhodopseudomonas palustris CGA009 | | Descriptor: | Alpha/beta hydrolase, DODECAETHYLENE GLYCOL | | Authors: | Nocek, B, Hajighasemi, M, Xu, X, Cui, H, Savchenko, A, Yakunin, A. | | Deposit date: | 2014-03-07 | | Release date: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of alpha/beta hydrolase from Rhodopseudomonas palustris CGA009
TO BE PUBLISHED
|
|
4IC1
 
 | | Crystal structure of SSO0001 | | Descriptor: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-09 | | Release date: | 2013-01-16 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
4MWA
 
 | | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Brunzelle, J.S, Xu, X, Cui, H, Maltseva, N, Bishop, B, Kwon, K, Savchenko, A, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
