1UKX
 
 | | Solution structure of the RWD domain of mouse GCN2 | | Descriptor: | GCN2 eIF2alpha kinase | | Authors: | Nameki, N, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RWD domain of the mouse GCN2 protein.
Protein Sci., 13, 2004
|
|
4JDP
 
 | | Crystal structure of probable p-nitrophenyl phosphatase (pho2) (target EFI-501307) from Archaeoglobus fulgidus DSM 4304 with Magnesium bound | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, p-nitrophenyl phosphatase (Pho2) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Allen, K.N, Dunaway-Mariano, D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of probable p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
1UDM
 
 | | Solution structure of Coactosin-like protein (Cofilin family) from Mus Musculus | | Descriptor: | Coactosin-like protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-01 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
1V6F
 
 | | Solution Structure of Glia Maturation Factor-beta from Mus Musculus | | Descriptor: | glia maturation factor, beta | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-29 | | Release date: | 2004-05-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
3FS4
 
 | | Crystal structure determination of Ostrich hemoglobin at 2.2 Angstrom resolution | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Sundaresan, S.S, Ramesh, P, Sivakumar, K, Ponnuswamy, M.N. | | Deposit date: | 2009-01-09 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural studies of hemoglobin from two flightless birds, ostrich and turkey: insights into their differing oxygen-binding properties.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4M1A
 
 | | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldella termitidis ATCC 33386 | | Descriptor: | Hypothetical protein | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldellatermitidis ATCC 33386
To be Published
|
|
3HRW
 
 | | Crystal structure of hemoglobin from mouse (Mus musculus)at 2.8 | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sundaresan, S.S, Ramesh, P, Ponnuswamy, M.N. | | Deposit date: | 2009-06-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of hemoglobin from mouse (Mus musculus) compared with those from other small animals and humans.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4QB5
 
 | | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118 | | Descriptor: | 1,2-ETHANEDIOL, Glyoxalase/bleomycin resistance protein/dioxygenase, SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-06 | | Release date: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118
To be Published
|
|
4MYM
 
 | | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, AL Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-27 | | Release date: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides.
To be Published
|
|
4J2U
 
 | | Crystal structure of an enoyl-CoA hydratase from Rhodobacter sphaeroides 2.4.1 | | Descriptor: | Enoyl-CoA hydratase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-05 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase from Rhodobacter sphaeroides 2.4.1
To be Published
|
|
4J3C
 
 | | Crystal structure of 16S ribosomal RNA methyltransferase RsmE | | Descriptor: | Ribosomal RNA small subunit methyltransferase E | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-05 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 16S ribosomal RNA methyltransferase RsmE
TO BE PUBLISHED
|
|
4JJ9
 
 | | Crystal Structure of 5-carboxymethyl-2-hydroxymuconate delta-isomerase | | Descriptor: | 5-carboxymethyl-2-hydroxymuconate delta-isomerase, SULFATE ION | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-07 | | Release date: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of 5-carboxymethyl-2-hydroxymuconate delta-isomerase
To be Published
|
|
4JOT
 
 | | Crystal structure of enoyl-CoA hydrotase from Deinococcus radiodurans R1 | | Descriptor: | Enoyl-CoA hydratase, putative, GLYCEROL | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-18 | | Release date: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of enoyl-CoA hydrotase from Deinococcus radiodurans R1
To be Published
|
|
4K2N
 
 | | Crystal structure of an enoyl-CoA hydratase/ carnithine racemase from Magnetospirillum magneticum | | Descriptor: | Enoyl-CoA hydratase/carnithine racemase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-09 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/ carnithine racemase from Magnetospirillum magneticum
To be Published
|
|
4JCS
 
 | | Crystal structure of Enoyl-CoA hydratase/isomerase from Cupriavidus metallidurans CH34 | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Eswaramoorthy, S, Chamala, S, Chamala, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Enoyl-CoA hydratase/isomerase from Cupriavidus metallidurans CH34
To be Published
|
|
4JFC
 
 | | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666 | | Descriptor: | Enoyl-CoA hydratase, GLYCEROL | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666
To be Published
|
|
4K3W
 
 | | Crystal structure of an enoyl-CoA hydratase/isomerase from Marinobacter aquaeolei | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-11 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/isomerase from Marinobacter aquaeolei
To be Published
|
|
4JCU
 
 | | Crystal structure of a 5-carboxymethyl-2-hydroxymuconate isomerase from Deinococcus radiodurans R1 | | Descriptor: | 5-carboxymethyl-2-hydroxymuconate isomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a 5-carboxymethyl-2-hydroxymuconate isomerase from Deinococcus radiodurans R1
To be Published
|
|
4K29
 
 | | Crystal structure of an enoyl-CoA hydratase/isomerase from Xanthobacter autotrophicus Py2 | | Descriptor: | Enoyl-CoA hydratase/isomerase, GLYCEROL, L(+)-TARTARIC ACID | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-08 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/isomerase from Xanthobacter autotrophicus Py2
TO BE PUBLISHED
|
|
4KPK
 
 | | Crystal structure of a enoyl-CoA hydratase from Shewanella pealeana ATCC 700345 | | Descriptor: | 1,2-ETHANEDIOL, Enoyl-CoA hydratase/isomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-13 | | Release date: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Shewanella pealeana ATCC 700345
To be Published
|
|
4LK5
 
 | | Crystal structure of a enoyl-CoA hydratase from Mycobacterium avium subsp. paratuberculosis K-10 | | Descriptor: | enoyl-CoA hydratase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-06 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Mycobacterium avium subsp. paratuberculosis K-10
To be Published
|
|
4KD6
 
 | | Crystal structure of a Enoyl-CoA hydratase/isomerase from Burkholderia graminis C4D1M | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a Enoyl-CoA hydratase/isomerase from Burkholderia graminis C4D1M
To be Published
|
|
4LKB
 
 | | Crystal structure of a putative 4-Oxalocrotonate Tautomerase from Nostoc sp. PCC 7120 | | Descriptor: | GLYCEROL, SULFATE ION, hypothetical protein alr4568/putative 4-Oxalocrotonate Tautomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-07 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a putative 4-Oxalocrotonate Tautomerase from Nostoc sp. PCC 7120
To be Published
|
|
1V5B
 
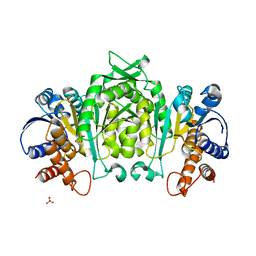 | | The Structure Of The Mutant, S225A and E251L, Of 3-Isopropylmalate Dehydrogenase From Bacillus Coagulans | | Descriptor: | 3-isopropylmalate dehydrogenase, SULFATE ION | | Authors: | Fujita, K, Minami, H, Suzuki, K, Tsunoda, M, Sekiguchi, T, Mizui, R, Tsuzaki, S, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a highly thermo-stabilized mutant of 3-isopropylmalate dehydrogenase from Bacillus coagulans: An evaluation of local packing density in the hydrophobic core
To be Published
|
|
1WFQ
 
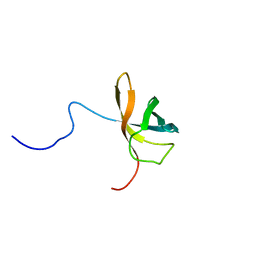 | | Solution structure of the first cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
