7DRI
 
 | | Structure of SspE_CTD_41658 | | Descriptor: | DUF1524 domain | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-28 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
4MZ4
 
 | | Discovery of an Irreversible HCV NS5B Polymerase Inhibitor | | Descriptor: | 1-[(2-chloroquinolin-3-yl)methyl]-6-fluoro-5-methyl-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Zeng, Q, Anilkumar, G.N, Rosenblum, S.B, Huang, H.-C, Lesburg, C.A, Jiang, Y, Selyutin, O, Chan, T.-Y, Bennett, F, Chen, K.X, Venkatraman, S, Sannigrahi, M, Velazquez, F, Duca, J.S, Gavalas, S, Huang, Y, Pu, H, Wang, L, Pinto, P, Vibulbhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Li, C, Hesk, D, Gesell, J, Sorota, S, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of an irreversible HCV NS5B polymerase inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5ZN8
 
 | | Crystal structure of nicotinamidase PncA from Bacillus subtilis | | Descriptor: | Isochorismatase, ZINC ION | | Authors: | Shang, F, Chen, J, Wang, L, Xu, Y. | | Deposit date: | 2018-04-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the nicotinamidase/pyrazinamidase PncA from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 503, 2018
|
|
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
3U4R
 
 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-N-({3-[(methylsulfonyl)amino]phenyl}sulfonyl)-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxamide, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannagrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3U1L
 
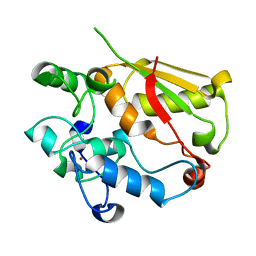 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
5Y9Q
 
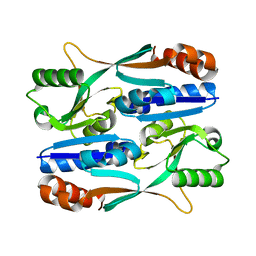 | |
3U4O
 
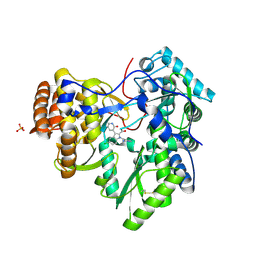 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannigrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6A2H
 
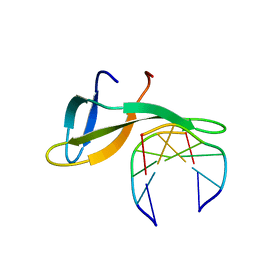 | | Architectural roles of Cren7 in folding crenarchaeal chromatin filament | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(P*AP*AP*TP*TP*AP*C)-3'), DNA (5'-D(P*GP*TP*AP*AP*TP*T)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Chen, Y.Y, Wang, L, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Mol. Microbiol., 111, 2019
|
|
7DRS
 
 | | Structure of SspE_40224 | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRR
 
 | | Structure of SspE-R100A protein | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
6A2I
 
 | | Architectural roles of Cren7 in folding crenarchaeal chromatin filament | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*CP*GP*TP*AP*GP*CP*TP*AP*AP*TP*TP*AP*GP*CP*TP*AP*CP*G)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Chen, Y.Y, Wang, L, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Mol. Microbiol., 111, 2019
|
|
6JBJ
 
 | | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family D member 4 | | Authors: | Xu, D, Feng, Z, Hou, W.T, Jiang, Y.L, Wang, L, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4.
Cell Res., 29, 2019
|
|
2ODA
 
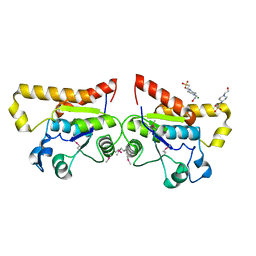 | | Crystal Structure of PSPTO_2114 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypothetical protein PSPTO_2114, MAGNESIUM ION | | Authors: | Peisach, E, Allen, K.N, Dunaway-Mariano, D, Wang, L, Burroughs, A.M, Aravind, L. | | Deposit date: | 2006-12-22 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray crystallographic structure and activity analysis of a Pseudomonas-specific subfamily of the HAD enzyme superfamily evidences a novel biochemical function.
Proteins, 70, 2007
|
|
8V6U
 
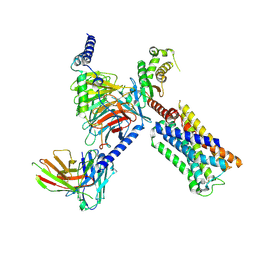 | | 5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking | | Descriptor: | 4-{(3R)-1-[(1R)-1-(pyrimidin-2-yl)ethyl]piperidin-3-yl}phenol, 5-hydroxytryptamine receptor 2A, G protein alpha-subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Gumpper, R.H, Wang, L, Kapolka, N, Skiniotis, G, Roth, B.L. | | Deposit date: | 2023-12-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
2MY9
 
 | |
7VFA
 
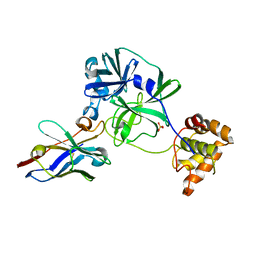 | | the complex of SARS-CoV2 3CL and NB1A2 | | Descriptor: | 3C-like proteinase, NB1A2, SULFATE ION | | Authors: | Sun, Z.C, Wang, L, Geng, Y. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VFB
 
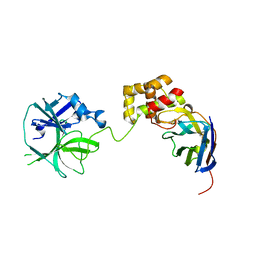 | | the complex of SARS-CoV2 3cl and NB2B4 | | Descriptor: | 3C-like proteinase, nb2b4 | | Authors: | Geng, Y, Sun, Z.C, Wang, L. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VWC
 
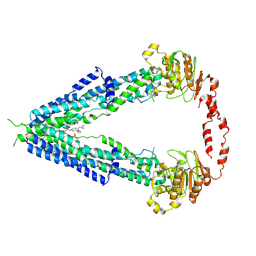 | | Cryo-EM structure of human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, [(2R)-3-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-2-oxidanyl-propyl] octadecanoate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-10 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
7VX8
 
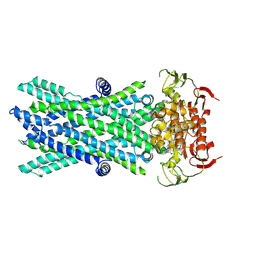 | | Cryo-EM structure of ATP-bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1 | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-12 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
7VZB
 
 | | Cryo-EM structure of C22:0-CoA bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] docosanethioate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
7XC6
 
 | |
2ZGJ
 
 | | Crystal Structure of D86N-GzmM Complexed with Its Optimal Synthesized Substrate | | Descriptor: | Granzyme M, SSGKVPLS, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-22 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
2ZGH
 
 | | Crystal Structure of active granzyme M bound to its product | | Descriptor: | Granzyme M, SSGKVPL, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-22 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
2ZKS
 
 | | Structural insights into the proteolytic machinery of apoptosis-inducing Granzyme M | | Descriptor: | Granzyme M, SULFATE ION, hGzmM inhibitor | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Yang, X, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-03-28 | | Release date: | 2009-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
