5P2P
 
 | | X-RAY STRUCTURE OF PHOSPHOLIPASE A2 COMPLEXED WITH A SUBSTRATE-DERIVED INHIBITOR | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2, PHOSPHONIC ACID 2-DODECANOYLAMINO-HEXYL ESTER PROPYL ESTER | | Authors: | Dijkstra, B.W, Thunnissen, M.M.G.M, Kalk, K.H, Drenth, J. | | Deposit date: | 1990-09-01 | | Release date: | 1991-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of phospholipase A2 complexed with a substrate-derived inhibitor.
Nature, 347, 1990
|
|
8BPP
 
 | |
8BPQ
 
 | |
8QQK
 
 | | Cryo-EM structure of E. coli cytochrome bo3 quinol oxidase assembled in peptidiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, ... | | Authors: | Gao, Y, Zhang, Y, Hakke, S, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2023-10-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of cytochrome bo 3 quinol oxidase assembled in peptidiscs reveals an "open" conformation for potential ubiquinone-8 release.
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
1ABQ
 
 | |
1ABO
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF THE ABL TYROSINE KINASE SH3 DOMAIN WITH 3BP-1 SYNTHETIC PEPTIDE | | Descriptor: | 3BP-1 SYNTHETIC PEPTIDE, 10 RESIDUES, ABL TYROSINE KINASE, ... | | Authors: | Musacchio, A, Wilmanns, M, Saraste, M. | | Deposit date: | 1995-05-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides.
Nat.Struct.Biol., 1, 1994
|
|
1AWO
 
 | |
5BYV
 
 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
1K5N
 
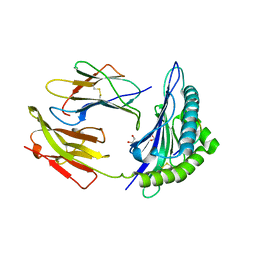 | | HLA-B*2709 BOUND TO NONA-PEPTIDE M9 | | Descriptor: | GLYCEROL, beta-2-microglobulin, light chain, ... | | Authors: | Hulsmeyer, M, Hillig, R.C, Volz, A, Ruhl, M, Schroder, W, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2001-10-11 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | HLA-B27 Subtypes Differentially Associated with Disease Exhibit Subtle Structural Alterations
J.Biol.Chem., 277, 2002
|
|
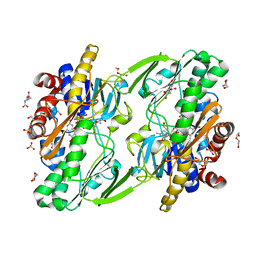
8DHK
 
 | |
8DYJ
 
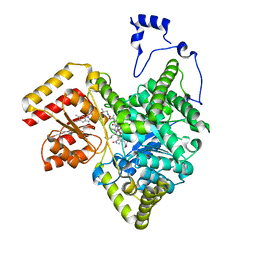 | | Crystal structure of human methylmalonyl-CoA mutase in complex with ADP and cob(II)alamin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COBALAMIN, Methylmalonyl-CoA mutase, ... | | Authors: | Mascarenhas, R.N, Gouda, H, Banerjee, R. | | Deposit date: | 2022-08-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bivalent molecular mimicry by ADP protects metal redox state and promotes coenzyme B 12 repair.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DYL
 
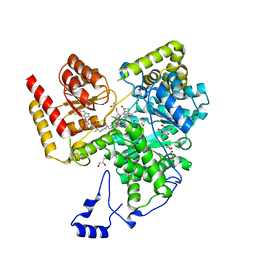 | | Crystal structure of human methylmalonyl-CoA mutase bound to aquocobalamin | | Descriptor: | COBALAMIN, GLYCEROL, Methylmalonyl-CoA mutase, ... | | Authors: | Mascarenhas, R.N, Gouda, H, Banerjee, R. | | Deposit date: | 2022-08-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bivalent molecular mimicry by ADP protects metal redox state and promotes coenzyme B 12 repair.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5KJK
 
 | | SMYD2 in complex with AZ370 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-[2-[1-[2-(3,4-dichlorophenyl)ethyl]azetidin-3-yl]oxyphenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)pyridine-4-carboxamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
5KJL
 
 | | SMYD2 in complex with AZ378 | | Descriptor: | N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
5KJN
 
 | | SMYD2 in complex with AZ506 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 5-[2-[4-[2-(1~{H}-indol-3-yl)ethyl]piperazin-1-yl]phenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)pyridine-3-carboxamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
5KJM
 
 | | SMYD2 in complex with AZ931 | | Descriptor: | 6-[2-[4-[2-(3,4-dichlorophenyl)ethyl]piperazin-1-yl]phenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxamide, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
7L7G
 
 | | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B from Homo sapiens (updated model of PDB ID: 6CAJ) | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Tsai, J.C, Miller-Vedam, L.E, Anand, A, Jaishankar, P, Nguyen, H.C, Wang, L, Renslo, A.R, Frost, A, Walter, P. | | Deposit date: | 2020-12-28 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | eIF2B conformation and assembly state regulates the integrated stress response.
Elife, 10, 2021
|
|
5P9G
 
 | | Structure of BTK with RN486 | | Descriptor: | 6-cyclopropyl-8-fluoranyl-2-[2-(hydroxymethyl)-3-[1-methyl-5-[[5-(4-methylpiperazin-1-yl)pyridin-2-yl]amino]-6-oxidanylidene-pyridin-3-yl]phenyl]isoquinolin-1-one, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5P9M
 
 | | BTK1 BINDS COVALENTLY TO HY-15771 ONO-4059 | | Descriptor: | 6-azanyl-9-[(3~{R})-1-[(~{E})-but-2-enoyl]pyrrolidin-3-yl]-7-(4-phenoxyphenyl)purin-8-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5P9K
 
 | | CRYSTAL STRUCTURE OF BTK with CNX 774 | | Descriptor: | 4-[4-[[5-fluoranyl-4-[[3-(propanoylamino)phenyl]amino]pyrimidin-2-yl]amino]phenoxy]-~{N}-methyl-pyridine-2-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5P9I
 
 | | BTK1 SOAKED WITH IBRUTINIB-Rev | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
7OLY
 
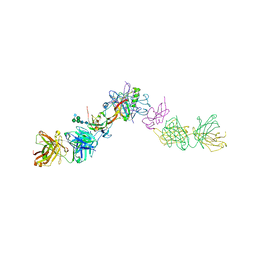 | | Structure of activin A in complex with an ActRIIB-Alk4 fusion reveal insight into activin receptor interactions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-1B, ... | | Authors: | Hakansson, M, Rose, N.C, Castonguay, R, Logan, D.T, Krishnan, L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.265 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
6MAC
 
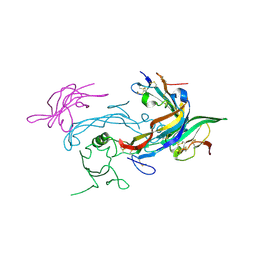 | | Ternary structure of GDF11 bound to ActRIIB-ECD and Alk5-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 11, ... | | Authors: | Goebel, E.J, Thompson, T.B. | | Deposit date: | 2018-08-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural characterization of an activin class ternary receptor complex reveals a third paradigm for receptor specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6N9P
 
 | |
