8RYZ
 
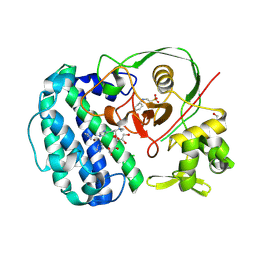 | | Structures of selenoneine synthase SenA from Variovorax paradoxus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, IMIDAZOLE, ... | | Authors: | Ma, Y.Y, Gao, Y, Xu, S.H. | | Deposit date: | 2024-02-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structures of SenB and SenA enzymes from Variovorax paradoxus provide insights into carbon-selenium bond formation in selenoneine biosynthesis.
Heliyon, 10, 2024
|
|
8XFB
 
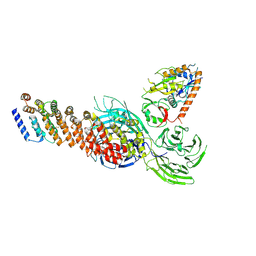 | | Cryo-EM structure of partial dimeric WDR11-FAM91A1 complex | | Descriptor: | Protein FAM91A1, WD repeat-containing protein 11 | | Authors: | Jia, G.W, Deng, Q.H, Su, Z.M, Jia, D. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins.
Cell, 187, 2024
|
|
7Y8B
 
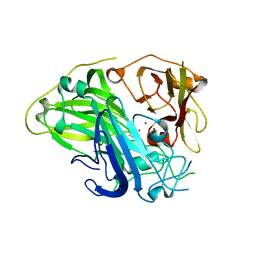 | | Crystal structure of CotA laccase complexed with syringic acid | | Descriptor: | 3,5-dimethoxy-4-oxidanyl-benzoic acid, COPPER (II) ION, Spore coat protein A, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2022-06-23 | | Release date: | 2023-06-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insights into substrate promiscuity of CotA laccase catalyzing lignin-phenol derivatives.
Int.J.Biol.Macromol., 256, 2023
|
|
7Y8C
 
 | | Crystal structure of CotA laccase complexed with syringaldehyde | | Descriptor: | 3,5-dimethoxy-4-oxidanyl-benzaldehyde, COPPER (II) ION, Spore coat protein A, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2022-06-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insights into substrate promiscuity of CotA laccase catalyzing lignin-phenol derivatives.
Int.J.Biol.Macromol., 256, 2023
|
|
8JYS
 
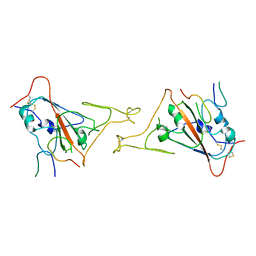 | |
7DAT
 
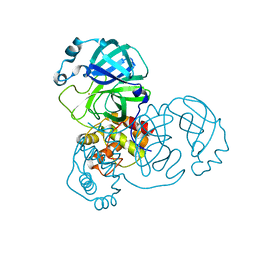 | | The crystal structure of COVID-19 main protease treated by AF | | Descriptor: | COVID-19 MAIN PROTEASE, GOLD ION | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7DAV
 
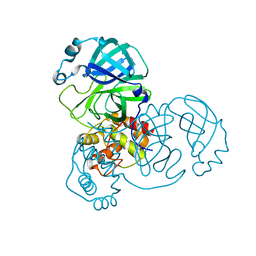 | | The native crystal structure of COVID-19 main protease | | Descriptor: | COVID-19 MAIN PROTEASE | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7DAU
 
 | | The crystal structure of COVID-19 main protease treated by GA | | Descriptor: | COVID-19 MAIN PROTEASE, GOLD ION | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7X83
 
 | | Cryo-EM structure of the TMEM106B fibril from normal elder | | Descriptor: | Transmembrane protein 106B | | Authors: | Xia, W.C, Zhao, Q.Y, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
7X84
 
 | | Cryo-EM structure of the TMEM106B fibril from Parkinson's disease dementia | | Descriptor: | Transmembrane protein 106B | | Authors: | Zhao, Q.Y, Xia, W.C, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
5YS1
 
 | | Crystal structure of Multicopper Oxidase CueO G304K mutant | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | Deposit date: | 2017-11-12 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
5YS5
 
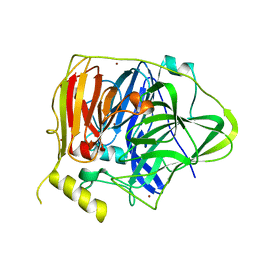 | | Crystal structure of Multicopper Oxidase CueO G304K mutant with seven copper ions | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | Deposit date: | 2017-11-13 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
8H92
 
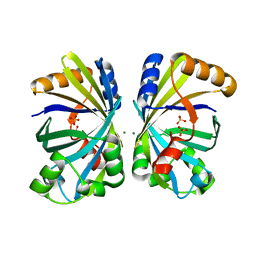 | |
7VH8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Zhao, Y, Zhang, Q, Yang, H, Rao, Z. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332.
Protein Cell, 13, 2022
|
|
6C10
 
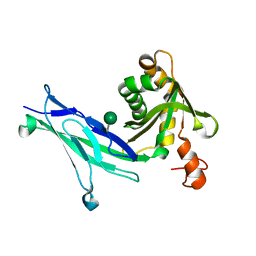 | | Crystal structure of mouse PCDH15 EC11-EL | | Descriptor: | Protocadherin-15, alpha-D-mannopyranose | | Authors: | Gouaux, E, Elferich, J, Ge, J. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5.
Elife, 7, 2018
|
|
6C14
 
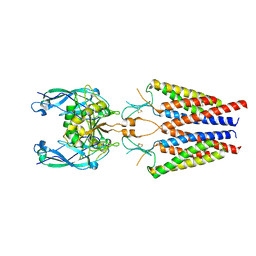 | |
6C13
 
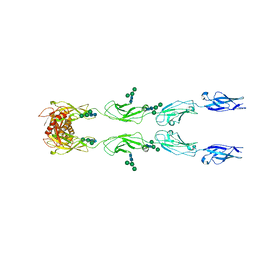 | | CryoEM structure of mouse PCDH15-4EC-LHFPL5 complex | | Descriptor: | Protocadherin-15, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gouaux, E, Ge, J, Elferich, J. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (11.33 Å) | | Cite: | Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5.
Elife, 7, 2018
|
|
4I8B
 
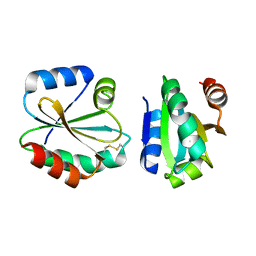 | | Crystal Structure of Thioredoxin from Schistosoma Japonicum | | Descriptor: | Thioredoxin | | Authors: | Wu, Q, Peng, Y, Zhao, J, Li, X, Fan, X, Zhou, X, Chen, J, Luo, Z, Shi, D. | | Deposit date: | 2012-12-03 | | Release date: | 2013-12-04 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression, characterization and crystal structure of thioredoxin from Schistosoma japonicum.
Parasitology, 142, 2015
|
|
7YQE
 
 | |
6BAW
 
 | | Structure of GRP94 with a selective resorcinylic inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, Endoplasmin, PHOSPHATE ION, ... | | Authors: | Que, N.L.S, Gewirth, D.T. | | Deposit date: | 2017-10-16 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure Based Design of a Grp94-Selective Inhibitor: Exploiting a Key Residue in Grp94 To Optimize Paralog-Selective Binding.
J. Med. Chem., 61, 2018
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
6CEO
 
 | |
