6RRY
 
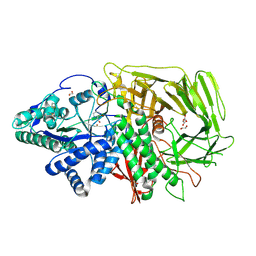 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3R)-2-(Hydroxymethyl)-1,2,3,6-tetrahydro-3-pyridinol | | Descriptor: | (2~{S},3~{R})-2-(hydroxymethyl)-1,2,3,6-tetrahydropyridin-3-ol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RQZ
 
 | | GOLGI ALPHA-MANNOSIDASE II complex with Manno-epi-cyclophellitol aziridine | | Descriptor: | (1~{R},2~{S},3~{R},4~{R},5~{S},6~{R})-5-azanyl-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, Debets, M, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RRW
 
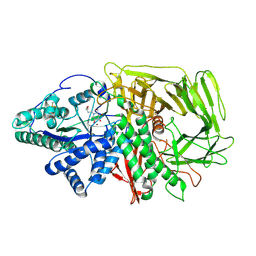 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2R,3R,4R,5S)-1-(5-{[4-(3,4-Dihydro-2H-1,5-benzodioxepin-7-yl)benzyl]oxy}pentyl)-2-(hydroxymethyl)-3,4,5-piperidinetriol | | Descriptor: | (2~{R},3~{R},4~{R},5~{S})-1-[5-[[4-(3,4-dihydro-2~{H}-1,5-benzodioxepin-7-yl)phenyl]methoxy]pentyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, Alpha-mannosidase 2, ZINC ION | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6MYL
 
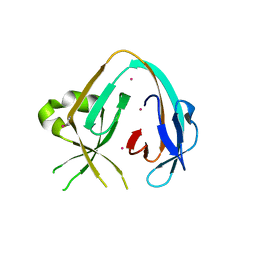 | | The Prp8 intein-cisplatin complex | | Descriptor: | PLATINUM (II) ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Li, H. | | Deposit date: | 2018-11-01 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Cisplatin protects mice from challenge ofCryptococcus neoformansby targeting the Prp8 intein.
Emerg Microbes Infect, 8, 2019
|
|
6LNZ
 
 | |
8QQI
 
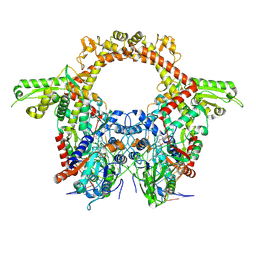 | | E.coli DNA gyrase in complex with 217 bp substrate DNA and LEI-800 | | Descriptor: | DNA gyrase subunit A, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Ghilarov, D, Martin, N.I, van der Stelt, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of isoquinoline sulfonamides as allosteric gyrase inhibitors with activity against fluoroquinolone-resistant bacteria.
Nat.Chem., 16, 2024
|
|
7X1M
 
 | | The complex structure of Omicron BA.1 RBD with BD604, S309,and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab heavy chain, BD-604 Fab light chain, ... | | Authors: | Huang, M, Xie, Y.F, Qi, J.X. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.
Immunity, 55, 2022
|
|
7XML
 
 | | Cryo-EM structure of PEIP-Bs_enolase complex | | Descriptor: | Enolase, MAGNESIUM ION, Putative gene 60 protein | | Authors: | Li, S, Zhang, K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage protein PEIP is a potent Bacillus subtilis enolase inhibitor.
Cell Rep, 40, 2022
|
|
8TLH
 
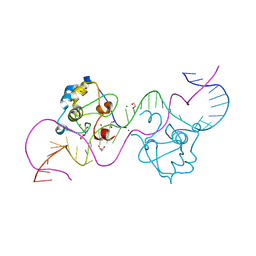 | | CDCA7 (Mouse) Binds Non-B-form 32-mer DNA oligo | | Descriptor: | 1,2-ETHANEDIOL, Cell division cycle-associated protein 7, DNA (32-MER), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The ICF syndrome protein CDCA7 harbors a unique DNA-binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Biorxiv, 2023
|
|
8TLF
 
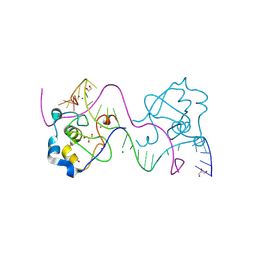 | | CDCA7 (Mouse) Binds Non-B-form DNA oligo 36-mer (sg C2-Form 2) | | Descriptor: | 1,2-ETHANEDIOL, Cell division cycle-associated protein 7, GLYCEROL, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The ICF syndrome protein CDCA7 harbors a unique DNA-binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Biorxiv, 2023
|
|
8TLK
 
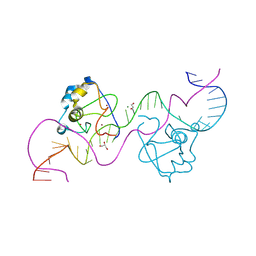 | | CDCA7 (Human) Binds Non-B-form 32-mer DNA oligo Containing a 5mC | | Descriptor: | 1,2-ETHANEDIOL, Cell division cycle-associated protein 7, DNA (32-MER), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ICF syndrome protein CDCA7 harbors a unique DNA-binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Biorxiv, 2023
|
|
8TLJ
 
 | | CDCA7 (Mouse) Binds Non-B-form 32-mer DNA oligo Containing a 5mC | | Descriptor: | 1,2-ETHANEDIOL, Cell division cycle-associated protein 7, DNA (32-MER), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The ICF syndrome protein CDCA7 harbors a unique DNA-binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Biorxiv, 2023
|
|
8TLL
 
 | | CDCA7 (Mouse) Binds Non-B-form 26-mer DNA oligo | | Descriptor: | 1,2-ETHANEDIOL, Cell division cycle-associated protein 7, DNA (26-MER), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The ICF syndrome protein CDCA7 harbors a unique DNA binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Sci Adv, 10, 2024
|
|
8TLE
 
 | |
8TLG
 
 | | CDCA7 (Mouse) Binds Non-B-form 34-mer DNA oligo | | Descriptor: | 1,2-ETHANEDIOL, Cell division cycle-associated protein 7, DNA (34-MER), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The ICF syndrome protein CDCA7 harbors a unique DNA-binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Biorxiv, 2023
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
1HVJ
 
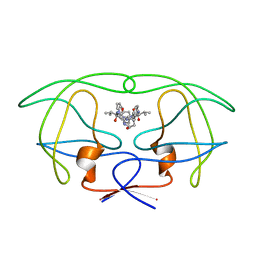 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-3-HYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
1HVK
 
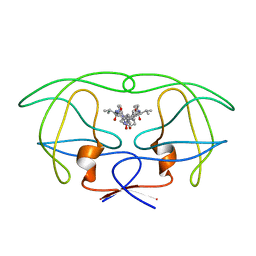 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2S,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
1HVI
 
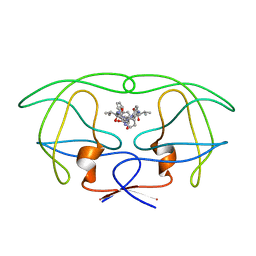 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2R,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
1HVL
 
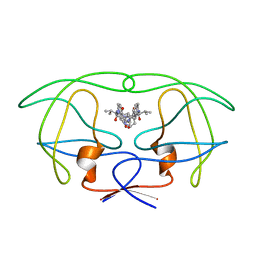 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2R,3R)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
8D6E
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with RP-6306 | | Descriptor: | (1P)-2-amino-1-(3-hydroxy-2,6-dimethylphenyl)-5,6-dimethyl-1H-pyrrolo[2,3-b]pyridine-3-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6D
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 39 | | Descriptor: | (1P)-2-amino-5-bromo-1-(3-hydroxy-2,6-dimethylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6F
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with Eph receptor inhibitor / compound 41 | | Descriptor: | (1M)-2-amino-1-(5-hydroxy-2-methylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, SULFATE ION | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
