1GSE
 
 | |
1GSF
 
 | | GLUTATHIONE TRANSFERASE A1-1 COMPLEXED WITH ETHACRYNIC ACID | | Descriptor: | ETHACRYNIC ACID, GLUTATHIONE TRANSFERASE A1-1 | | Authors: | L'Hermite, G, Sinning, I, Cameron, A.D, Jones, T.A. | | Deposit date: | 1995-06-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of human alpha-class glutathione transferase A1-1 in the apo-form and in complexes with ethacrynic acid and its glutathione conjugate.
Structure, 3, 1995
|
|
6BXY
 
 | | Menin in complex with MI-1481 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-1-{[(2S)-5-oxomorpholin-2-yl]methyl}-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Borkin, D, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-19 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Complexity of Blocking Bivalent Protein-Protein Interactions: Development of a Highly Potent Inhibitor of the Menin-Mixed-Lineage Leukemia Interaction.
J.Med.Chem., 61, 2018
|
|
1GSD
 
 | |
5D6S
 
 | | Structure of epoxyqueuosine reductase from Streptococcus thermophilus. | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, IRON/SULFUR CLUSTER | | Authors: | Payne, K.A.P, Fisher, K, Dunstan, M.S, Sjuts, H, Leys, D. | | Deposit date: | 2015-08-12 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Epoxyqueuosine Reductase Structure Suggests a Mechanism for Cobalamin-dependent tRNA Modification.
J.Biol.Chem., 290, 2015
|
|
1GNE
 
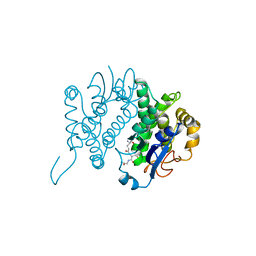 | | THE THREE-DIMENSIONAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE OF SCHISTOSOMA JAPONICUM FUSED WITH A CONSERVED NEUTRALIZING EPITOPE ON GP41 OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Lim, K, Ho, J.X, Keeling, K, Gilliland, G.L, Ji, X, Ruker, F, Carter, D.C. | | Deposit date: | 1994-06-16 | | Release date: | 1994-11-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV.
Protein Sci., 3, 1994
|
|
5D29
 
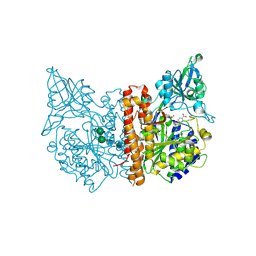 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a hydroxamate inhibitor JHU241 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(2~{S})-2-carboxy-5-(oxidanylamino)-5-oxidanylidene-pentyl]benzoic acid, ... | | Authors: | Barinka, C, Novakova, Z, Pavlicek, J. | | Deposit date: | 2015-08-05 | | Release date: | 2016-04-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unprecedented Binding Mode of Hydroxamate-Based Inhibitors of Glutamate Carboxypeptidase II: Structural Characterization and Biological Activity.
J.Med.Chem., 59, 2016
|
|
3NAX
 
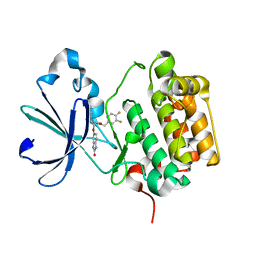 | | PDK1 in complex with inhibitor MP7 | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and pharmacological inhibition of PDK1 in cancer cells: characterization of a selective allosteric kinase inhibitor.
J.Biol.Chem., 286, 2011
|
|
8R56
 
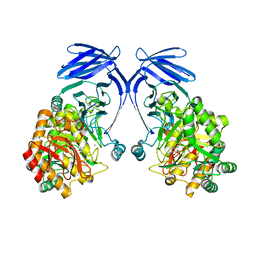 | |
8PYX
 
 | |
8PPP
 
 | |
8PYY
 
 | |
3NAY
 
 | | PDK1 in complex with inhibitor MP6 | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 4-(2-cyclopropylethylidene)-9-(1H-pyrazol-4-yl)-6-{[(1R)-1,2,2-trimethylpropyl]amino}benzo[c][1,6]naphthyridin-1(4H)-one | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective inhibition of PDK1 using an allosteric kinase inhibitor and RNAi impairs cancer cell migration and anchorage-independent growth of primary tumor lines
J.Biol.Chem., 2010
|
|
5ELY
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a hydroxamate inhibitor JHU242 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(2~{R})-2-carboxy-5-(oxidanylamino)-5-oxidanylidene-pentyl]benzoic acid, ... | | Authors: | Barinka, C, Novakova, Z, Pavlicek, J. | | Deposit date: | 2015-11-05 | | Release date: | 2016-04-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Unprecedented Binding Mode of Hydroxamate-Based Inhibitors of Glutamate Carboxypeptidase II: Structural Characterization and Biological Activity.
J.Med.Chem., 59, 2016
|
|
7ZNM
 
 | | Artificial Unspecific Peroxygenase expressed in Pichia pastoris at 2.01 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Artificial Unspecific Peroxygenase, ... | | Authors: | Robinson, W.X.Q, Mielke, T, Grogan, G. | | Deposit date: | 2022-04-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Comparing the Catalytic and Structural Characteristics of a 'Short' Unspecific Peroxygenase (UPO) Expressed in Pichia pastoris and Escherichia coli.
Chembiochem, 24, 2023
|
|
7ZNV
 
 | | Artificial Unspecific Peroxygenase expressed in Pichia pastoris at 1.21 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Robinson, W.X.Q, Mielke, T, Grogan, G. | | Deposit date: | 2022-04-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Comparing the Catalytic and Structural Characteristics of a 'Short' Unspecific Peroxygenase (UPO) Expressed in Pichia pastoris and Escherichia coli.
Chembiochem, 24, 2023
|
|
7ZNW
 
 | |
1FRO
 
 | |
4MVA
 
 | | 1.43 Angstrom Resolution Crystal Structure of Triosephosphate Isomerase (tpiA) from Escherichia coli in Complex with Acetyl Phosphate. | | Descriptor: | 1,2-ETHANEDIOL, ACETYLPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Kuhn, M.L, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
2CBO
 
 | | Crystal structure of the neocarzinostatin 3Tes24 mutant bound to testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, SULFATE ION, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2CBT
 
 | | Crystal structure of the neocarzinostatin 4Tes1 mutant bound testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2CBM
 
 | | Crystal structure of the apo-form of a neocarzinostatin mutant evolved to bind testosterone. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NEOCARZINOSTATIN | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
1GOZ
 
 | |
1GUH
 
 | | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the MU and PI class enzymes | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1-1, S-BENZYL-GLUTATHIONE | | Authors: | Sinning, I, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 1993-02-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes.
J.Mol.Biol., 232, 1993
|
|
2CBQ
 
 | | Crystal structure of the neocarzinostatin 1Tes15 mutant bound to testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, SULFATE ION, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
