1CTS
 
 | |
6JHU
 
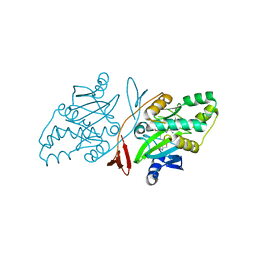 | | Crystal Structure Of Biotin Protein Ligase From Leishmania Major in complex with Biotinyl-5-AMP | | Descriptor: | BIOTINYL-5-AMP, Biotin/lipoate protein ligase-like protein, SULFATE ION | | Authors: | Rajak, M, Patel, A, Sundd, M. | | Deposit date: | 2019-02-19 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Leishmania major biotin protein ligase forms a unique cross-handshake dimer
Acta Crystallogr.,Sect.D, 77, 2021
|
|
5YO2
 
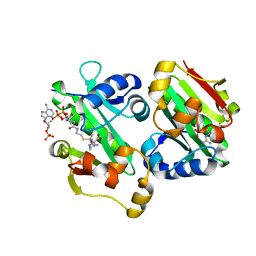 | | The crystal structure of Rv2747 from Mycobacterium tuberculosis in complex with Acetyl CoA and L-Arginine | | Descriptor: | ACETYL COENZYME *A, ARGININE, Amino-acid acetyltransferase | | Authors: | Singh, E, Tiruttani Subhramanyam, U.K, Pal, R.K, Srinivasan, A, Gourinath, S, Das, U. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural insights into the substrate binding mechanism of novel ArgA from Mycobacterium tuberculosis
Int. J. Biol. Macromol., 125, 2019
|
|
3CSC
 
 | |
5ZAU
 
 | |
3CTS
 
 | |
6ADD
 
 | | The crystal structure of Rv2747 from Mycobacterium tuberculosis in complex with CoA and NLQ | | Descriptor: | Amino-acid acetyltransferase, COENZYME A, N~2~-ACETYL-L-GLUTAMINE | | Authors: | Das, U, Singh, E, Pal, R.K, Tiruttani Subhramanyam, U.K, Gourinath, S, Srinivasan, A. | | Deposit date: | 2018-07-31 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural insights into the substrate binding mechanism of novel ArgA from Mycobacterium tuberculosis
Int. J. Biol. Macromol., 125, 2019
|
|
5YD2
 
 | |
5YII
 
 | |
3UAS
 
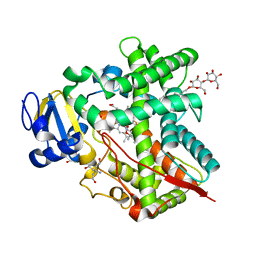 | | Cytochrome P450 2B4 covalently bound to the mechanism-based inactivator 9-ethynylphenanthrene | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gay, S.C, Zhang, H, Shah, M.B, Stout, C.D, Halpert, J.R, Hollenberg, P.F. | | Deposit date: | 2011-10-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | Potent Mechanism-Based Inactivation of Cytochrome P450 2B4 by 9-Ethynylphenanthrene: Implications for Allosteric Modulation of Cytochrome P450 Catalysis.
Biochemistry, 52, 2013
|
|
8TDO
 
 | |
8TDN
 
 | |
8TGO
 
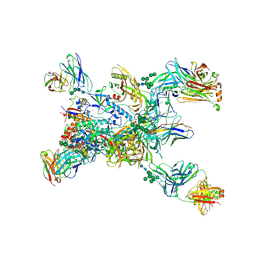 | | Crystal structure of the BG505 triple tandem trimer gp140 HIV-1 Env in complex with PGT124 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 scFv, ... | | Authors: | Xian, Y, Yuan, M, Wilson, I.A. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines.
Npj Vaccines, 9, 2024
|
|
5YB0
 
 | |
3TK3
 
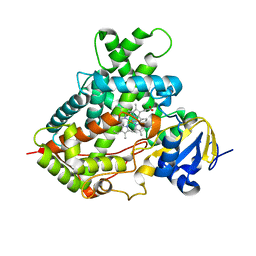 | | Cytochrome P450 2B4 mutant L437A in complex with 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Jang, H.H, Wilderman, P.R, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-08-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active-site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme.
Febs J., 279, 2012
|
|
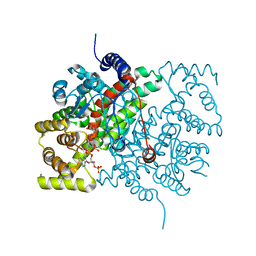
5CTS
 
 | |
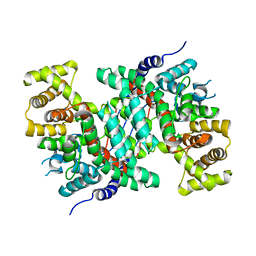
5CSC
 
 | |
6AVN
 
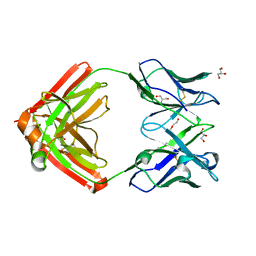 | |
6B0N
 
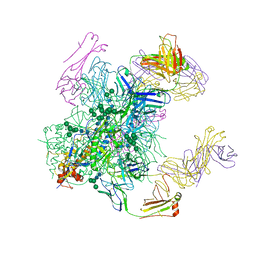 | | Crystal structure of the cleavage-independent prefusion HIV Env glycoprotein trimer of the clade A BG505 isolate (NFL construct) in complex with Fabs PGT122 and PGV19 at 3.39 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp140, ... | | Authors: | Sarkar, A, Irimia, A, Wilson, I.A. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer.
Nat Commun, 9, 2018
|
|
1OUY
 
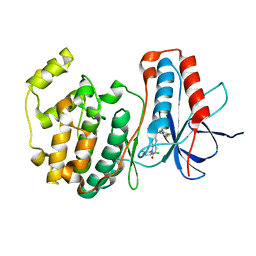 | | The structure of p38 alpha in complex with a dihydropyrido-pyrimidine inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-6-[(2,4-DIFLUOROPHENYL)SULFANYL]-7-(1,2,3,6-TETRAHYDRO-4-PYRIDINYL)-3,4-DIHYDROPYRIDO[3,2-D]PYRIMIDIN-2(1H)-ONE, Mitogen-activated protein kinase 14 | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
4LGX
 
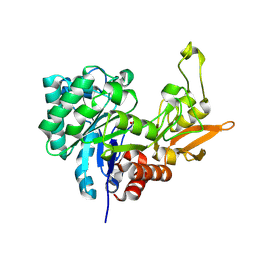 | | Structure of Chitinase D from Serratia proteamaculans revealed an unusually constrained substrate binding site | | Descriptor: | ACETATE ION, GLYCEROL, Glycoside hydrolase family 18 | | Authors: | Madhuprakash, J, Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2013-06-30 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Inverse relationship between chitobiase and transglycosylation activities of chitinase-D from Serratia proteamaculans revealed by mutational and biophysical analyses.
Sci Rep, 5, 2015
|
|
2CSC
 
 | |
2CTS
 
 | |
1ERI
 
 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | Authors: | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | Deposit date: | 1994-05-18 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
8GBL
 
 | | Structure of Apo Human SIRT5 | | Descriptor: | NAD-dependent protein deacylase sirtuin-5, mitochondrial, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
