3HF3
 
 | | Old Yellow Enzyme from Thermus scotoductus SA-01 | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, SULFATE ION | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
2R8X
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CHLORIDE ION | | Authors: | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | Deposit date: | 2007-09-11 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
4CF5
 
 | |
1A2S
 
 | | THE SOLUTION NMR STRUCTURE OF OXIDIZED CYTOCHROME C6 FROM THE GREEN ALGA MONORAPHIDIUM BRAUNII, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Banci, L, Bertini, I, De La Rosa, M.A, Koulougliotis, D, Navarro, J.A, Walter, O. | | Deposit date: | 1998-01-10 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized cytochrome c6 from the green alga Monoraphidium braunii.
Biochemistry, 37, 1998
|
|
4CDG
 
 | | Crystal structure of the Bloom's syndrome helicase BLM in complex with Nanobody | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BLOOM SYNDROME PROTEIN, NANOBODY, ... | | Authors: | Newman, J.A, Savitsky, P, Allerston, C.K, Pike, A.C.W, Pardon, E, Steyaert, J, Arrowsmith, C.H, von Delft, F, Bountra, C, Edwards, A, Gileadi, O. | | Deposit date: | 2013-10-31 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structure of the Bloom'S Syndrome Helicase Indicates a Role for the Hrdc Domain in Conformational Changes.
Nucleic Acids Res., 43, 2015
|
|
4CF3
 
 | |
4CE6
 
 | |
1WO6
 
 | | Solution structure of Designed Functional Finger 5 (DFF5): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
154D
 
 | | DNA DISTORTION IN BIS-INTERCALATED COMPLEXES | | Descriptor: | BIS-(N-ETHYLPYRIDINIUM-(3-METHOXYCARBAZOLE))HEXANE-1,6-DIAMINE, DNA (5'-D(*(CBR)P*GP*CP*G)-3') | | Authors: | Peek, M.E, Lipscomb, L.A, Bertrand, J.A, Gao, Q, Roques, B.P, Garbay-Jaureguiberry, C, Williams, L.D. | | Deposit date: | 1993-12-14 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA distortion in bis-intercalated complexes.
Biochemistry, 33, 1994
|
|
1M4C
 
 | | Crystal Structure of Human Interleukin-2 | | Descriptor: | interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4C9I
 
 | | Crystal Structure of the Strawberry Pathogenesis-Related 10 (PR-10) Fra a 1E protein (Form B) | | Descriptor: | (2R,3S)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol, GLYCEROL, MAJOR STRAWBERRY ALLERGEN FRA A 1-E | | Authors: | Casanal, A, Zander, U, Valpuesta, V, Marquez, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Strawberry Pathogenesis-Related 10 (Pr-10) Fra a Proteins Control Flavonoid Biosynthesis by Binding to Metabolic Intermediates.
J.Biol.Chem., 288, 2013
|
|
1WVN
 
 | |
1MOE
 
 | | The three-dimensional structure of an engineered scFv T84.66 dimer or diabody in VL to VH linkage. | | Descriptor: | SULFATE ION, anti-CEA mAb T84.66 | | Authors: | Carmichael, J.A, Power, B.E, Garrett, T.P.J, Yazaki, P.J, Shively, J.E, Raubischek, A.A, Wu, A.M, Hudson, P.J. | | Deposit date: | 2002-09-09 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of an Anti-CEA scFv Diabody Assembled from T84.66 scFvs in VL-to-VH Orientation: Implications for Diabody Flexibility
J.Mol.Biol., 326, 2003
|
|
4C8Y
 
 | |
1M4A
 
 | | Crystal Structure of Human Interleukin-2 Y31C Covalently Modified at C31 with (1H-Indol-3-yl)-(2-mercapto-ethoxyimino)-acetic acid | | Descriptor: | (1H-INDOL-3-YL)-(2-MERCAPTO-ETHOXYIMINO)-ACETIC ACID, GLYCEROL, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M49
 
 | | Crystal Structure of Human Interleukin-2 Complexed with SP-1985 | | Descriptor: | 2-[2-(1-CARBAMIMIDOYL-PIPERIDIN-3-YL)-ACETYLAMINO]-3-{4-[2-(3-OXALYL-1H-INDOL-7-YL)ETHYL]-PHENYL}-PROPIONIC ACID METHYL ESTER, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1A6Z
 
 | | HFE (HUMAN) HEMOCHROMATOSIS PROTEIN | | Descriptor: | BETA-2-MICROGLOBULIN, HFE | | Authors: | Lebron, J.A, Bennett, M.J, Vaughn, D.E, Chirino, A.J, Snow, P.M, Mintier, G.A, Feder, J.N, Bjorkman, P.J. | | Deposit date: | 1998-03-04 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor.
Cell(Cambridge,Mass.), 93, 1998
|
|
1MTP
 
 | | The X-ray crystal structure of a serpin from a thermophilic prokaryote | | Descriptor: | Serine Proteinase Inhibitor (SERPIN), Chain A, Chain B | | Authors: | Irving, J.A, Cabrita, L.D, Rossjohn, J, Pike, R.N, Bottomley, S.P, Whisstock, J.C. | | Deposit date: | 2002-09-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A crystal structure of a prokaryote serpin: controlling conformational change in a heated environment
Structure, 11, 2003
|
|
1WEG
 
 | | Catalytic Domain Of Muty From Escherichia Coli K142A Mutant | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, IMIDAZOLE, ... | | Authors: | Hitomi, K, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
1MUD
 
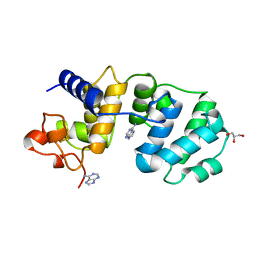 | | CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI, D138N MUTANT COMPLEXED TO ADENINE | | Descriptor: | ADENINE, Adenine DNA glycosylase, GLYCEROL, ... | | Authors: | Guan, Y, Tainer, J.A. | | Deposit date: | 1998-08-20 | | Release date: | 1999-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily.
Nat.Struct.Biol., 5, 1998
|
|
2R8Y
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli in a complex with Ca | | Descriptor: | CALCIUM ION, CHLORIDE ION, YrbI from Escherichia coli | | Authors: | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | Deposit date: | 2007-09-11 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
1WO4
 
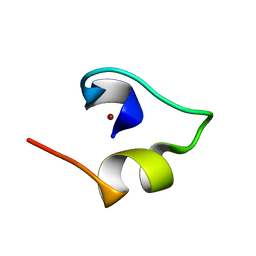 | | Solution structure of Minimal Mutant 2 (MM2): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
4CSZ
 
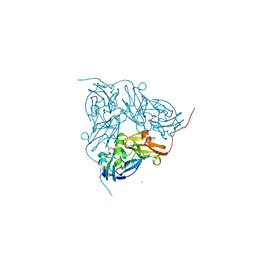 | | STRUCTURE OF F306C MUTANT OF NITRITE REDUCTASE FROM Achromobacter XYLOSOXIDANS WITH NITRITE BOUND | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Leferink, N.G.H, Antonyuk, S.V, Houwman, J.A, Scrutton, N.S, REady, R, Hasnain, S.S. | | Deposit date: | 2014-03-11 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Impact of Residues Remote from the Catalytic Centre on Enzyme Catalysis of Copper Nitrite Reductase.
Nat.Commun., 5, 2014
|
|
4CH2
 
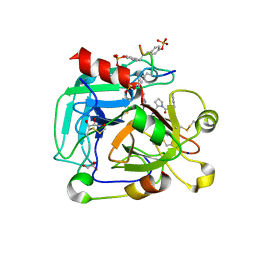 | | Low-salt crystal structure of a thrombin-GpIbalpha peptide complex | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, PLATELET GLYCOPROTEIN IB ALPHA CHAIN, ... | | Authors: | Lechtenberg, B.C, Freund, S.M.V, Huntington, J.A. | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gpibalpha Interacts Exclusively with Exosite II of Thrombin
J.Mol.Biol., 426, 2014
|
|
1MUO
 
 | | CRYSTAL STRUCTURE OF AURORA-2, AN ONCOGENIC SERINE-THREONINE KINASE | | Descriptor: | ADENOSINE, Aurora-related kinase 1 | | Authors: | Cheetham, G.M.T, Knegtel, R.M.A, Coll, J.T, Renwick, S.B, Swenson, L, Weber, P, Lippke, J.A, Austen, D.A. | | Deposit date: | 2002-09-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Aurora-2, an Oncogenic Serine/Threonine Kinase
J.Biol.Chem., 277, 2002
|
|
