7JIV
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
5XOB
 
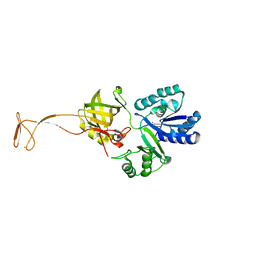 | | Crystal structure of apo TiaS (tRNAIle2 agmatidine synthetase) | | Descriptor: | MAGNESIUM ION, ZINC ION, tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS | | Authors: | Dong, J. | | Deposit date: | 2017-05-27 | | Release date: | 2018-08-29 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of tRNA-Modifying Enzyme TiaS and Motions of Its Substrate Binding Zinc Ribbon.
J. Mol. Biol., 430, 2018
|
|
6JCA
 
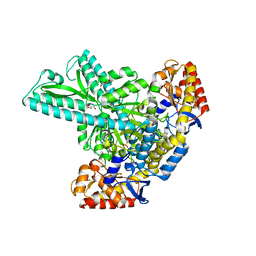 | |
5XVM
 
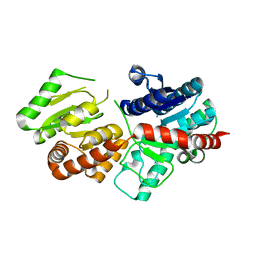 | |
4NSW
 
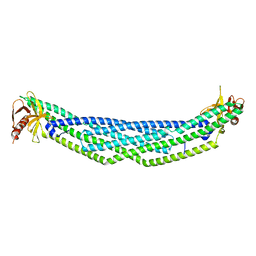 | | Crystal structure of the BAR-PH domain of ACAP1 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | Authors: | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | Deposit date: | 2013-11-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A PH Domain in ACAP1 Possesses Key Features of the BAR Domain in Promoting Membrane Curvature
Dev.Cell, 31, 2014
|
|
5XVJ
 
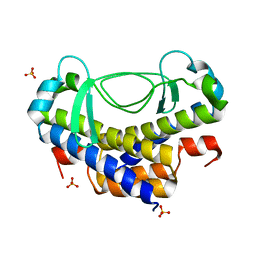 | | Crystal structure of AL7 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 7, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
5Z5K
 
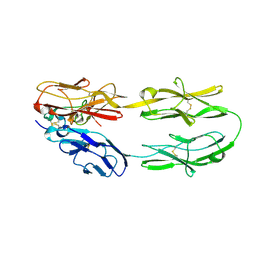 | | Structure of the DCC-Draxin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Draxin, ... | | Authors: | Liu, Y, Xiao, J, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2018-06-20 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural Basis for Draxin-Modulated Axon Guidance and Fasciculation by Netrin-1 through DCC.
Neuron, 97, 2018
|
|
7YHS
 
 | | Structure of Csy-AcrIF4-dsDNA | | Descriptor: | AcrIF4, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Feng, Y, Zhang, L.X. | | Deposit date: | 2022-07-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Anti-CRISPR protein AcrIF4 inhibits the type I-F CRISPR-Cas surveillance complex by blocking nuclease recruitment and DNA cleavage.
J.Biol.Chem., 298, 2022
|
|
7DRI
 
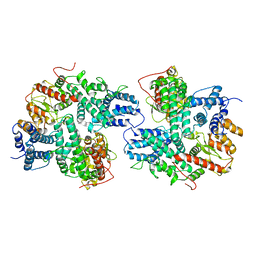 | | Structure of SspE_CTD_41658 | | Descriptor: | DUF1524 domain | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-28 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRS
 
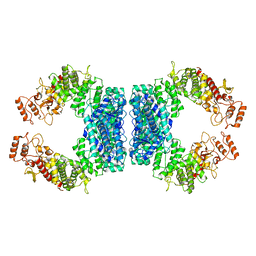 | | Structure of SspE_40224 | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRR
 
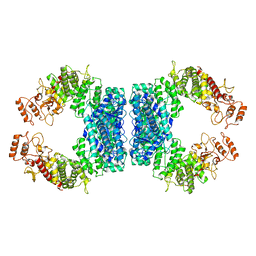 | | Structure of SspE-R100A protein | | Descriptor: | SspE protein | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
5GIY
 
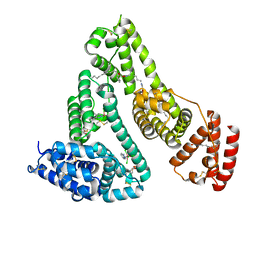 | | HSA-Palmitic acid-[RuCl5(ind)]2- | | Descriptor: | PALMITIC ACID, Serum albumin, pentakis(chloranyl)-(1~{H}-indazol-2-ium-2-yl)ruthenium(1-) | | Authors: | Yang, F, Wang, T. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Structure of HSA-Palmitic acid-[RuCl5(ind)]2-
To Be Published
|
|
5XVL
 
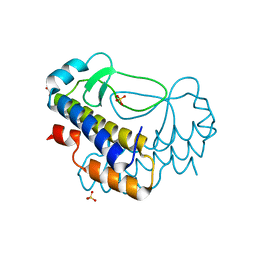 | | Crystal structure of AL2 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 2, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
6JC8
 
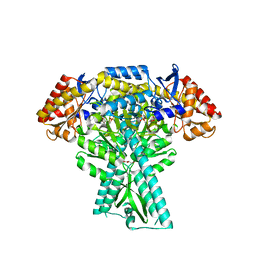 | |
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
5ZTF
 
 | | Structure of Ca2+ ATPase | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Inoue, M, Watanabe, S, Inaba, K. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Basis of Sarco/Endoplasmic Reticulum Ca2+-ATPase 2b Regulation via Transmembrane Helix Interplay.
Cell Rep, 27, 2019
|
|
6J75
 
 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase | | Descriptor: | Aldehyde dehydrogenase A | | Authors: | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-01-17 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
7XQ2
 
 | | Structure of hSLC19A1+2'3'-cGAMP | | Descriptor: | Reduced folate transporter, cGAMP | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ1
 
 | | Structure of hSLC19A1+2'3'-CDAS | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XPZ
 
 | | Structure of Apo-hSLC19A1 | | Descriptor: | Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ0
 
 | | Structure of hSLC19A1+3'3'-CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
6KX2
 
 | | Crystal structure of GDP bound RhoA protein | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
7X6Z
 
 | |
7X70
 
 | |
