5B2S
 
 | | Crystal structure of the Streptococcus pyogenes Cas9 EQR variant in complex with sgRNA and target DNA (TGAG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
5B2R
 
 | | Crystal structure of the Streptococcus pyogenes Cas9 VQR variant in complex with sgRNA and target DNA (TGA PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
3WND
 
 | | Crystal structure of EF-Pyl | | Descriptor: | CITRIC ACID, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
5H0Q
 
 | | Crystal structure of lipid binding protein Nakanori at 1.5A | | Descriptor: | Lipid binding protein | | Authors: | Makino, A, Abe, M, Ishitsuka, R, Murate, M, Kishimoto, T, Sakai, S, Hullin-Matsuda, F, Shimada, Y, Inaba, T, Miyatake, H, Tanaka, H, Kurahashi, A, Pack, C.G, Kasai, R.S, Kubo, S, Schieber, N.L, Dohmae, N, Tochio, N, Hagiwara, K, Sasaki, Y, Aida, Y, Fujimori, F, Kigawa, T, Nishikori, K, Parton, R.G, Kusumi, A, Sako, Y, Anderluh, G, Yamashita, M, Kobayashi, T, Greimel, P, Kobayashi, T. | | Deposit date: | 2016-10-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C
FASEB J., 31, 2017
|
|
3WS4
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2A) | | Descriptor: | Beta-lactamase, CHLORIDE ION, STRONTIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WRT
 
 | | Wild type beta-lactamase DERIVED FROM CHROMOHALOBACTER SP.560 | | Descriptor: | Beta-lactamase | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS2
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1C) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS1
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6M04
 
 | | Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8D | | Authors: | Nakamura, R, Kasuya, G, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation.
Commun Biol, 3, 2020
|
|
1EIE
 
 | |
1EKG
 
 | | MATURE HUMAN FRATAXIN | | Descriptor: | FRATAXIN | | Authors: | Dhe-Paganon, S, Shigeta, R, Chi, Y.I, Ristow, M, Shoelson, S.E. | | Deposit date: | 2000-03-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human frataxin.
J.Biol.Chem., 275, 2000
|
|
1EID
 
 | |
1EIC
 
 | |
4U4V
 
 | | Structure of a nitrate/nitrite antiporter NarK in apo inward-open state | | Descriptor: | NICKEL (II) ION, Nitrate/nitrite transporter NarK, OLEIC ACID | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4U4W
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
1FS3
 
 | |
4U4T
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound inward-open state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4UDD
 
 | | GR in complex with desisobutyrylciclesonide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DESISOBUYTYRYL CICLESONIDE, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDC
 
 | | GR in complex with dexamethasone | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DEXAMETHASONE, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDA
 
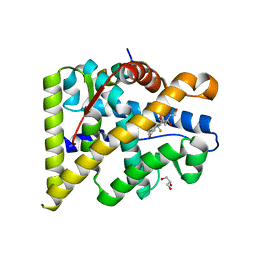 | | MR in complex with dexamethasone | | Descriptor: | DEXAMETHASONE, GLYCEROL, MINERALOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
5AYN
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POTASSIUM ION, Solute carrier family 39 (Iron-regulated transporter) | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
5AYO
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in inward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POTASSIUM ION, Solute carrier family 39 (Iron-regulated transporter), ... | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
5AYM
 
 | | Crystal structure of a bacterial homologue of iron transporter ferroportin in outward-facing state with soaked iron | | Descriptor: | FE (II) ION, Solute carrier family 39 (Iron-regulated transporter) | | Authors: | Taniguchi, R, Kato, H.E, Font, J, Deshpande, C.N, Ishitani, R, Jormakka, M, Nureki, O. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin
Nat Commun, 6, 2015
|
|
8WG3
 
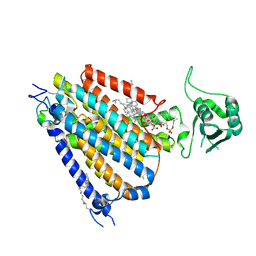 | | mouse TMEM63b in LMNG-CHS micelle | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
8WG4
 
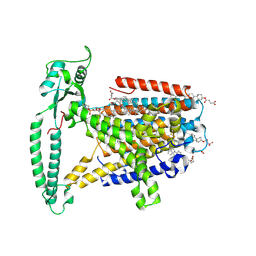 | | mouse TMEM63b in DDM-CHS micelle with YN9303-24 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
