5WYX
 
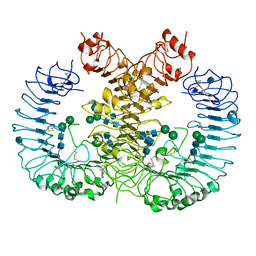 | | Crystal structure of human TLR8 in complex with CU-CPT8m | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(3-methylphenyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small-molecule inhibition of TLR8 through stabilization of its resting state
Nat. Chem. Biol., 14, 2018
|
|
4YT0
 
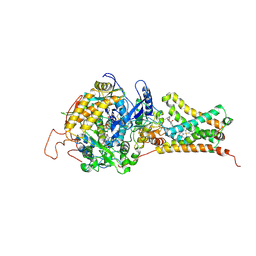 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide. | | Descriptor: | 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
7EQ9
 
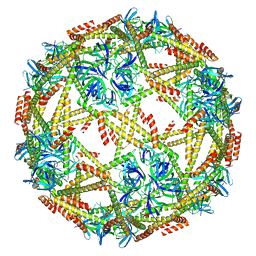 | | Cryo-EM structure of designed protein nanoparticle TIP60 (Truncated Icosahedral Protein composed of 60-mer fusion proteins) | | Descriptor: | TIP60 | | Authors: | Obata, J, Kawakami, N, Tsutsumi, A, Miyamoto, K, Kikkawa, M, Arai, R. | | Deposit date: | 2021-04-30 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Icosahedral 60-meric porous structure of designed supramolecular protein nanoparticle TIP60.
Chem.Commun.(Camb.), 57, 2021
|
|
4YSY
 
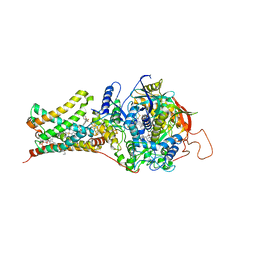 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-[(2,4-dichlorophenyl)methyl]-2-(trifluoromethyl)benzamide | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
1IYL
 
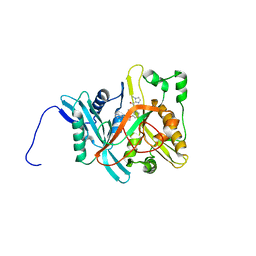 | | Crystal Structure of Candida albicans N-myristoyltransferase with Non-peptidic Inhibitor | | Descriptor: | (1-METHYL-1H-IMIDAZOL-2-YL)-(3-METHYL-4-{3-[(PYRIDIN-3-YLMETHYL)-AMINO]-PROPOXY}-BENZOFURAN-2-YL)-METHANONE, Myristoyl-CoA:Protein N-Myristoyltransferase | | Authors: | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
5B7V
 
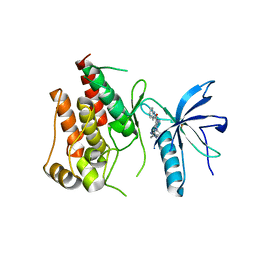 | | Human FGFR1 kinase in complex with CH5183284 | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, [5-amino-1-(2-methyl-1H-benzimidazol-6-yl)-1H-pyrazol-4-yl](1H-indol-2-yl)methanone | | Authors: | Fukami, T.A, Lukacs, C.M, Janson, C. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor
Mol.Cancer Ther., 13, 2014
|
|
1IYK
 
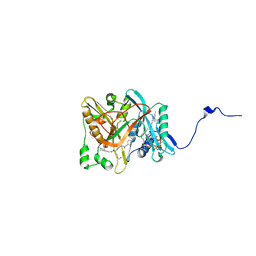 | | Crystal structure of candida albicans N-myristoyltransferase with myristoyl-COA and peptidic inhibitor | | Descriptor: | MYRISTOYL-COA:PROTEIN N-MYRISTOYLTRANSFERASE, TETRADECANOYL-COA, [CYCLOHEXYLETHYL]-[[[[4-[2-METHYL-1-IMIDAZOLYL-BUTYL]PHENYL]ACETYL]-SERYL]-LYSINYL]-AMINE | | Authors: | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
3VA2
 
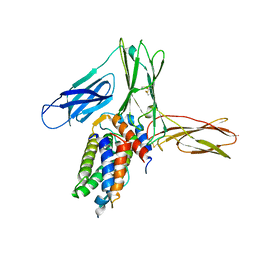 | | Crystal structure of human Interleukin-5 in complex with its alpha receptor | | Descriptor: | Interleukin-5, Interleukin-5 receptor subunit alpha | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-12-28 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural basis of interleukin-5 dimer recognition by its alpha receptor
Protein Sci., 21, 2012
|
|
2ZXQ
 
 | | Crystal structure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Endo-alpha-N-acetylgalactosaminidase, MANGANESE (II) ION | | Authors: | Suzuki, R, Katayama, T, Ashida, H, Yamamoto, K, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational analyses of substrate recognition of endo-{alpha}-N-acetylgalactosaminidase from Bifidobacterium longum.
J.Biochem., 2009
|
|
2KF3
 
 | |
1NCP
 
 | |
8WX2
 
 | |
8WX5
 
 | |
8W42
 
 | | X-ray crystal structure of V30M-TTR in complex with resveratrol | | Descriptor: | RESVERATROL, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W44
 
 | | X-ray crystal structure of V30M-TTR in complex with oxyresveratrol | | Descriptor: | SODIUM ION, Transthyretin, trans-oxyresveratrol | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W46
 
 | | X-ray crystal structure of V30M-TTR in complex with pterostilbene | | Descriptor: | Pterostilbene, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W43
 
 | | X-ray crystal structure of V30M-TTR in complex with piceatannol | | Descriptor: | PICEATANNOL, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W45
 
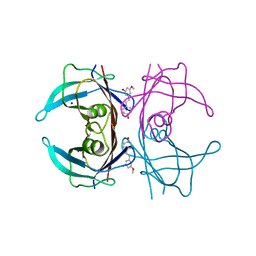 | | X-ray crystal structure of V30M-TTR in complex with pinostilbene | | Descriptor: | 3-[(E)-2-(4-hydroxyphenyl)ethenyl]-5-methoxy-phenol, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8W47
 
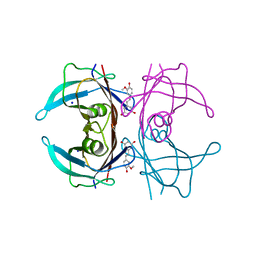 | | X-ray crystal structure of V30M-TTR in complex with isorhapontigenin | | Descriptor: | 5-[(~{E})-2-(3-methoxy-4-oxidanyl-phenyl)ethenyl]benzene-1,3-diol, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
8WX1
 
 | |
2KF6
 
 | |
2KF4
 
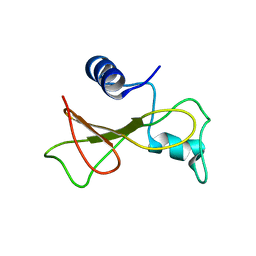 | | Barnase high pressure structure | | Descriptor: | Ribonuclease | | Authors: | Williamson, M.P, Wilton, D.J. | | Deposit date: | 2009-02-11 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Pressure-dependent structure changes in barnase on ligand binding reveal intermediate rate fluctuations.
Biophys.J., 97, 2009
|
|
2KF5
 
 | |
3VJK
 
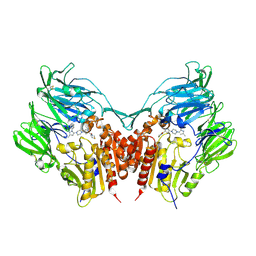 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with MP-513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
5Y6Q
 
 | |
