1V0D
 
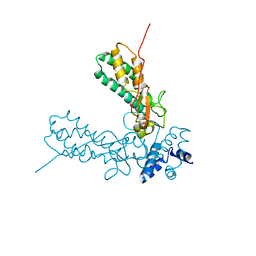 | | Crystal Structure of Caspase-activated DNase (CAD) | | Descriptor: | DNA FRAGMENTATION FACTOR 40 KDA SUBUNIT, LEAD (II) ION, MAGNESIUM ION, ... | | Authors: | Woo, E.-J, Kim, Y.-G, Kim, M.-S, Han, W.-D, Shin, S, Oh, B.-H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Mechanism for Inactivation and Activation of Cad/Dff40 in the Apoptotic Pathway
Mol.Cell, 14, 2004
|
|
3H77
 
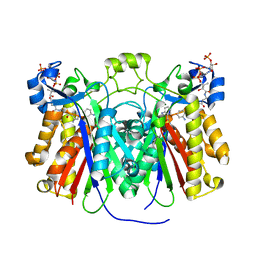 | | Crystal structure of Pseudomonas aeruginosa PqsD in a covalent complex with anthranilate | | Descriptor: | Anthraniloyl-coenzyme A, PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3H78
 
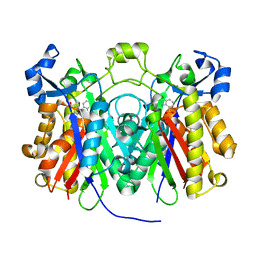 | | Crystal structure of Pseudomonas aeruginosa PqsD C112A mutant in complex with anthranilic acid | | Descriptor: | 2-AMINOBENZOIC ACID, PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3CAA
 
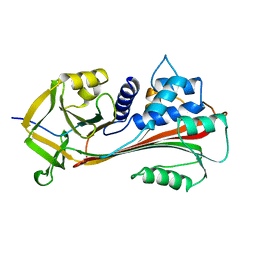 | | CLEAVED ANTICHYMOTRYPSIN A347R | | Descriptor: | ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
3H76
 
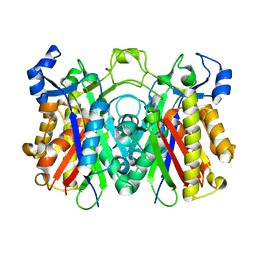 | | Crystal structure of PqsD, a key enzyme in Pseudomonas aeruginosa quinolone signal biosynthesis pathway | | Descriptor: | PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3B2R
 
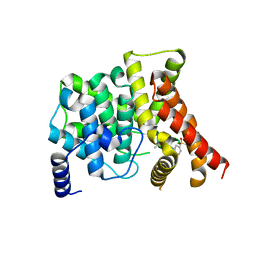 | | Crystal Structure of PDE5A1 catalytic domain in complex with Vardenafil | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Huanchen, W, Mengchun, Y, Howard, R, Sharron, H.F, Hengming, K. | | Deposit date: | 2007-10-19 | | Release date: | 2008-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Conformational variations of both phosphodiesterase-5 and inhibitors provide the structural basis for the physiological effects of vardenafil and sildenafil.
Mol.Pharmacol., 73, 2008
|
|
3BJC
 
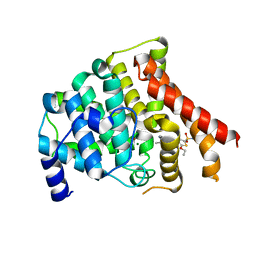 | | Crystal structure of the PDE5A catalytic domain in complex with a novel inhibitor | | Descriptor: | 5-ethoxy-4-(1-methyl-7-oxo-3-propyl-6,7-dihydro-1H-pyrazolo[4,3-d]pyrimidin-5-yl)thiophene-2-sulfonamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, G, Wang, H, Howard, R, Cai, J, Wan, Y, Ke, H. | | Deposit date: | 2007-12-03 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An insight into the pharmacophores of phosphodiesterase-5 inhibitors from synthetic and crystal structural studies
BIOCHEM.PHARM., 75, 2008
|
|
3HGU
 
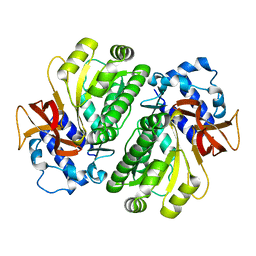 | |
3HGV
 
 | |
3FD9
 
 | |
2HTH
 
 | | Structural basis for ubiquitin recognition by the human EAP45/ESCRT-II GLUE domain | | Descriptor: | Ubiquitin, Vacuolar protein sorting protein 36 | | Authors: | Alam, S.L, Whitby, F.G, Hill, C.P, Sundquist, W.I. | | Deposit date: | 2006-07-25 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3ZXP
 
 | |
1AS4
 
 | | CLEAVED ANTICHYMOTRYPSIN A349R | | Descriptor: | ACETATE ION, ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
1Z7Q
 
 | | Crystal structure of the 20s proteasome from yeast in complex with the proteasome activator PA26 from Trypanosome brucei at 3.2 angstroms resolution | | Descriptor: | Potential proteasome component C5, Proteasome component C1, Proteasome component C11, ... | | Authors: | Forster, A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2005-03-26 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
2QYM
 
 | | crystal structure of unliganded PDE4C2 | | Descriptor: | MAGNESIUM ION, Phosphodiesterase 4C, ZINC ION | | Authors: | Ke, H. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
2QYL
 
 | | Crystal structure of PDE4B2B in complex with inhibitor NPV | | Descriptor: | 4-[8-(3-nitrophenyl)-1,7-naphthyridin-6-yl]benzoic acid, MAGNESIUM ION, Phosphodiesterase 4B, ... | | Authors: | Ke, H. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
2QYN
 
 | | Crystal structure of PDE4D2 in complex with inhibitor NPV | | Descriptor: | 4-[8-(3-nitrophenyl)-1,7-naphthyridin-6-yl]benzoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ke, H. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
3T5S
 
 | |
4A2U
 
 | | CRP(CAP) from Myco. Tuberculosis, with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY CRP/ FNR-FAMILY) | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2011-09-28 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | A New Crystal Form of Crp from M. Tuberculosis and Conformational Effects of Crystal Packing
To be Published
|
|
1RCW
 
 | |
1OMO
 
 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
3V4U
 
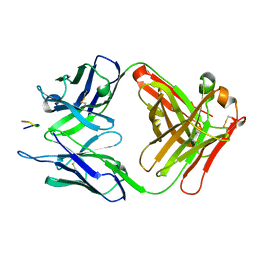 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, 64-3-7 L CHAIN, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-12-15 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
3UYR
 
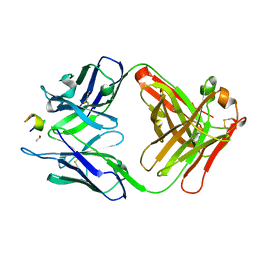 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | 1,2-ETHANEDIOL, H-2 class I histocompatibility antigen, L-D alpha chain, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-12-06 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
3UO1
 
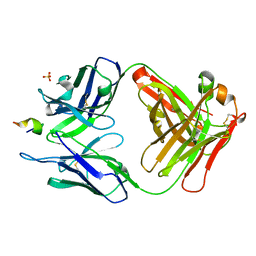 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, 64-3-7 L CHAIN, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-11-16 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
1O06
 
 | | Crystal structure of the Vps27p Ubiquitin Interacting Motif (UIM) | | Descriptor: | Vacuolar protein sorting-associated protein VPS27, ZINC ION | | Authors: | Fisher, R.D, Wang, B, Alam, S.L, Higginson, D.S, Rich, R, Myszka, D, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and ubiquitin binding of the ubiquitin-interacting motif.
J.Biol.Chem., 278, 2003
|
|
